Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast
|
|
|
- Rudolph Small
- 6 years ago
- Views:
Transcription
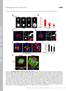
1 Current Biology, Volume 23 Supplemental Information Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast Felipe O. Bendezú and Sophie G. Martin 1
2 2
3 Figure S1 (Related to Figure 1). Cells Committed to a Mate Can Re-enter Exploratory Phase and Engage with Another Mate; Detail of Dynamic Exploration (A) DIC and GFP fluorescence images of h90 cells encoding a GFP fusion to the M factor receptor Map3. Note expression of Map3-GFP in h+ cells marked with dark blue circles at 4.7 hour time point. Light blue circles mark h- cells that do not expresses Map3-GFP. Dashed white lines indicate sister cells and red arrows indicate mated pairs. Time is in hours. (B) DIC images of wildtype h90 mating cells. The cell marked with an arrowhead shows a visible shmoo towards a prospective mate. When the prospective mate fuses with another cell the marked cell stops shmooing (arrowhead) and later engages with another mate (arrow). Time is in hours. (C and D) DIC (upper) and GFP fluorescence (lower) images of wildtype h90 scd1-3gfp mating cells. Arrowheads show small shmoo growth that stops when prospective mate chooses another partner. Arrows show Scd1 re-enter exploratory phase. Time is in minutes. (E) Deconvolved maximum-intensity projection of 3D DeltaVision imaging of Scd1-3GFP signals in h90 mating cells showing zones (arrowhead) and dots (arrow) in an exploring cell and single zones (red arrowhead) with no dots in engaged cells. Dot signals are weak and not detected by either OAI imaging or spinning disk confocal microscopy shown in the other Figures. Time is in minutes. Sum = max-intensity projection of 25 images over 2 hours. (F) OAI imaging of Scd1-3GFP and Scd2-mCherry signals in heterothallic h+ and h- mating cells, respectively. Arrowheads indicate coincident zones between cells of distinct mating type. DIC image shows fusion of cells following partner choice. The high percentage of zones localized adjacent to a possible partner (left) indicates zone location is influenced by partner position. Bars are 5 µm. 3
4 4
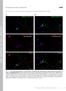
5 Figure S2 (Related to Figure 2). Exocyst Member Sec8 Explores Cell Periphery, but Cell Wall Synthases Bgs1 and Bgs4 Localize to Internal Structures during Cdc42 Exploration (A) OAI imaging showing exploration of Sec8-GFP. (B) Middle optical z slice of the same cells shown in Figure 2C. Arrowheads indicate dynamic Scd2-mCherry zones. (C) Absence of colocalization of Bgs4-GFP with Scd2-mCherry exploratory zones. Bgs4 localizes to distinct internal structures, which are often adjacent but not overlapping with Scd2 signal at the periphery. Right panels show the middle optical z slice only for indicated time points. (D) Colocalization of Bgs4-GFP and Scd2-mCherry at shmoo tips (red arrowhead). (E) Staining of h90 exploring cells with Cell Mask, which labels the plasma membrane. This shows localization of Scd1 zones at the cell periphery (arrowheads). In contrast, Bgs1 label internal structures that do not colocalize with Cell Mask during exploration (arrow), while these enzymes localize at the cell periphery at shmoo tips (arrowheads). 5
6 6
7 Figure S3 (Related to Figure 3). Scd1-3GFP Zone Dynamics Quantification after Short and Prolonged Exposure to P Factor, Exploratory Polarization in pka1 Cells, and Mating- Type Switching Rates in Hypersignaling Mutants (A) Quantification of Scd1-3GFP zones per cell of h- sxa2 cells treated with methanol (MeOH) solvent control or increasing concentration of P-factor. Cells were grown in MSL-N for 4 hours before placement onto MSL-N agarose pads and incubated at 25 o C for 2 or 14 hours before imaging. Cells were imaged by OAI microscopy every 5 minutes for 1 hour. % side indicates the percent of zones present along the cell sides. Sides were defined as the cylindrical part of the cell, and comprise an estimated 33% of total surface area. (B) Scd1-3GFP signals in h- sxa2 pka1 cells imaged on MSL media containing nitrogen and either P-factor or solvent methanol. We note cells treated with methanol were actively growing and dividing, indicating presence of sufficient nutrient for growth, whereas the majority of cells treated with P-factor had either arrested growth (0.1µg/ml) as judged by smaller cell size or were actively shmooing (1µg/ml). Arrowheads show zones of Scd1. Bar is 5 µm. (C) Homothallic h90 cells switch mating types in a pattern such that half the cells in a population are born with a compatible sister and the other half are born with a sister of the same mating type. The percent of cell pairs born with compatible sisters in wt h90, h90 map3 dn9 -GFP and h90 rgs1 was determined by imaging the mating-type specific expression of the pheromone receptor Map3-GFP or the truncated Map3 dn9 -GFP fusions. 7
8 Table S1. Strains Used in This Study YSM1623 h90 bgs1::ura4+pbgs1-gfp-bgs1-leu1+ leu1-32 ura4-d18 This work YSM2038 h90 sec8-gfp-ura4+ This work YSM2039 h+ scd1-3gfp-kanmx This work YSM2040 h90 scd1-3gfp-kanmx This work YSM2041 h90 scd1-3gfp-kanmx scd2-mcherry-natmx This work YSM2042 h- scd2-mcherry-natmx This work YSM2043 h90 scd1-3gfp-kanmx myo52-dttomato-natmx This work YSM2044 h- sxa2::kanmx scd1-3gfp-kanmx This work YSM2045 h90 map3::ura4+ Pmap3-map3 dn9 -GFP-kan This work YSM2046 h90 map3::ura4+ Pmap3-map3 dn9 -GFP-kan scd2-mcherry-nat This work YSM2047 h90 sec6-gfp-ura4+ scd2-mcherry-natmx This work This work YSM2048 h90 bgs4::ura4+pbgs4-gfp-bgs4-leu1+ scd2-mcherrynatmx YSM2049 h90 scd1-3gfp-hph rgs1::kanmx This work YSM2050 h90 scd2-mcherry-natmx ura4-294::pshk1-scgic2-crib- This work 3GFP-ura4+ YSM2122 h90 ura4-294::pshk1-scgic2-crib-3gfp-ura4+ This work This work YSM2123 h90 bgs1::ura4+pbgs1-gfp-bgs1-leu1+ scd2-mcherrynatmx YSM2124 h- sxa2::kanmx scd1-3gfp-kanmx pka1::hphmx This work YSM2125 h90 map3::ura4+ Pmap3-map3-GFP-kan This work YSM2126 h90 map3::ura4+ Pmap3-map3-GFP-kan rgs1::hphmx This work 8
9 Supplemental Experimental Procedures Strains, Media, and Growth Conditions S. pombe strains used are listed in Supplementary Table 1. Cells were grown in MSL +N or MSL N (minimal sporulation liquid or agar) [33]. MSL N solid media was used as either 2% agar plates or 2% electrophoresis-grade agarose pads. Agarose pads were covered with a coverslip and sealed with VALAP (1:1:1 Vaseline:Lanolin:Paraffin). Strains containing sxa2::kanmx and rgs1::kanmx mutations were obtained from the Bioneer haploid mutant collection and verified by diagnostic PCR. For construction of scd2-mcherry-natmx6, a PCR-based approach was used with plasmid pfa6a-mcherry-natmx6 [34] and oligos with homology to 78 nt directly upstream and 78 nt downstream the stop codon. Gene disruptions and tagging were confirmed by diagnostic PCR for both sides of the gene replacements or insertions. P-factor pheromone was purchased from Pepnome (Zhuhai City, China) and used from a stock solution of 1 mg/ml in methanol. CellMask orange plasma membrane strain (C10045) was purchased from Invitrogen and added to mating cells at a final concentration of 2 µg/ml for 10 minutes followed by a single wash with MSL-N before imaging. Mating Assays Cells were grown in pre-culture in MSL +N at 25ºC to OD 600 =0.4 to 1 and diluted to OD 600 =0.025 in MSL +N (for heterothallic mixtures in Fig. S1F cells were mixed in equal parts). Cells were then grown for hours to OD 600 =0.4 to 1 at either 25 or 30ºC in MSL+N. Cells were harvested by centrifugation and washed 3 times in MSL N. From here, cells were treated in one of three different ways depending on the experiment. For most fluorescent imaging (Fig. 1D-G, Fig. 2A-D, Fig. 3C-D and Fig. S1B-E, Fig. S2A-E), cells were placed onto MSL N plates and incubated for 18 hours at 18 o C. Cells were then resuspended in MSL N liquid and placed onto MSL N pad for imaging. For DIC imaging of pre-arrested cells or CRIB-GFP imaging (Fig.1A-C, Fig. 3E-F and Fig.S1A), cells were resuspended to OD 600 =20 and 0.3 µl placed onto MSL N pads. For heterothallic cells in Fig. S1F, cells were resuspended to OD 600 =1.5 and allowed to arrest in G1 at 30 o C for 4 hours and placed onto MSL N pad. We note that CRIB-GFP expression did not seem to have an effect on mating properties: h90 CRIB-GFP cells mated with similar kinetics and efficiency as wildtype cells. We further note that Scd2 dynamics observed in CRIB-GFP-expressing cells was indistinguishable from that of wildtype cells. Pheromone Treatment Heterothallic cells were grown in pre-culture in MSL +N at 25 o C to OD 600 =0.4 to 1 and diluted to OD 600 =0.025 in MSL +N. Cells were then grown for 18 hours to OD 600 =0.4 to 1 at 30 o C. Cells were washed 3 times in MSL N and resuspended to OD 600 =1.5 and allowed to arrest in G1 at 30 o C for 4 hours and placed onto MSL N pad (Fig. 3A) or 35mm petri plate (Fig. 3B) with methanol or P-factor. For cells treated with additional P-factor, 2 µl of P-factor (or methanol as control) was added around all four sides of cover slip and incubated for an additional 18 hours. For imaging of Scd1-3GFP in pka1 (Fig. S3B), cells were grown in MSL+N at 25 o C to OD 600 = and placed directly onto MSL+N pads with methanol or P-factor and incubated 14 hours at 18 o C before imaging. 9
10 Microscopy and Image Analysis Images were acquired on either a spinning disk confocal microscope or a DeltaVision epifluorescence system. The DeltaVision platform (Applied Precision) was composed of a customized Olympus IX-71 inverted microscope and a Plan Apo 60 /1.42 NA (for DIC) or a U- Plan Apo 100 /1.4 NA oil objective (for fluorescence), a CoolSNAP HQ2 camera (Photometrics), and an Insight SSI 7 color combined unit illuminator. To limit photobleaching, images in Fig. 1D, F-G, Fig. 3A, and Fig. S1C-D, Fig. S1F and Fig. S2A were captured by OAI (optical axis integration) imaging of a 4.6 µm z section, which is essentially a real time z sweep. High-resolution imaging was performed with either the Delta Vision or spinning disk confocal microscope. For Delta Vision imaging, optical z slices were captured every 0.3 µm (Fig. S1E) or every 0.6 µm (Fig. 3B-D) and maximum projections are shown. Images were acquired with softworx software. The spinning disk microscope system was as previously described [35]. For spinning disk confocal imaging (Fig. 1C and E, Fig. 2A-D, Fig. S2B-E and Fig. S3B) optical slices were acquired every 0.6 µm and maximum projections are shown for all except for Fig. S2B and E and the right panels of C where the medial optical slice is shown. Control experiments confirmed co-localization was not the result of optical bleed-through between the green and red channels by imaging single color fluorescent strains. All imaging was performed at room temperature (20-22 o C). Peripheral kymographs were constructed in ImageJ v1.46 by drawing a 5 pixel (0.65 µm) line around the cell cortex. Zone properties (Fig. 1G) were calculated from 20 kymographs by placing oval ROIs over areas visually deemed a zone. Quantification of shmoo position (Figure 1B, upper) was accomplished by determining the origin of each shmoo. Tip shmoos were those that originated from within the hemispherical cell tip and side shmoos if originated between the two hemispherical cell tips. Shmoo angles (Figure 1B, lower) were measured with the ObjectJ plugin for Image J by comparing the axis of shmoo growth to the axis of growth before arrest with 0 o indicating no change in direction and 90 o representing shmoo growth perpendicular to the growth axis before arrest. We note that for this analysis the cellular location of the shmoo is not included and could have originated from the cell tip or cell side. The distribution of shmoo angles in Fig. 1B and Fig. 3E are shown graphically by binning the angles into 15 o groups from 0 to 90 o. The length of the pie slice represents the percent of angles in each angle group normalized to the maximum. Figures were assembled with Adobe Photoshop CS5 and Adobe Illustrator CS5. Calculation of Engagement Frequencies Mating behavior frequencies were calculated as follows, with fused pairs (asci) and tetrads counting as two cells: Engagement frequency = (engaged cells)/((engaged cells) + (non-engaged shmooing cells) + (unmated cells)). Non-engaged shmooing cell frequency = (non-engaged shmooing cells)/((engaged cells) + (non-engaged shmooing cells) + (unmated cells)). 3 partners group frequency = (engaged triplets)/(engaged cells). Sister pair frequency = (engaged sisters)/(engaged cells). Determination of Mating-Type Switching Rates and Partner Choice of Compatible Sister Cells The rate of mating type switching in wt h90 map3-gfp, h90 map3 dn9 -GFP and h90 rgs1 map3- GFP strains was calculated by determining the percent of compatible sister pairs (one h+ and one h-) amongst all sister pairs. This was done by timelapse imaging of Map3( dn9 )-GFP 10
11 expression during growth arrest and the subsequent mating response on MSL-N agarose pads. As the Map3 M-factor receptor is only expressed in h+ cells, compatible sisters cells were determined as sister pairs where only one cell showed a GFP signal. For determining the partner choice of compatible sisters, the percent of sisters that chose another cell over a compatible sister was assessed. Sister pairs were included in the analysis only if of opposite mating types (as assessed from Map3-GFP expression) and in close proximity of at least one other compatible cell. Only the choice of the first sister cell to mate was considered, as the second cell to engage no longer has an available sister. Estimation of Cell Volume And Cell Surfaces Cell dimensions were measured from DIC images using the ObjectJ plugin for ImageJ. The volume of rod shaped and shmooing cells were calculated as a cylinder with two hemispherical caps where r is the radius and h is the height of a cylinder (total length - 2r): (4/3) r 3 + r 3 h. The surface of each hemispherical end is (4 r 2 )/2 and the surface of the cylinder is 2 rh. As starved, exploring cells are about 6 µm in length and 4 µm wide, cell side surface represents about 33% of the total surface. 11
12 Supplemental References 33. Egel, R., Willer, M., Kjaerulff, S., Davey, J., and Nielsen, O. (1994). Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast 10, Snaith, H.A., Samejima, I., and Sawin, K.E. (2005). Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. Embo J 24, Bendezú, F.O., Vincenzetti, V., and Martin, S.G. (2012). Fission yeast sec3 and exo70 are transported on actin cables and localize the exocyst complex to cell poles. PLoS One 7, e
Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion
 Supplemental data for: Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion Omaya Dudin, Laura Merlini and Sophie G Martin Figure S1 (related to Figure 1): Myo51 and
Supplemental data for: Spatial focalization of pheromone/mapk signaling triggers commitment to cell-cell fusion Omaya Dudin, Laura Merlini and Sophie G Martin Figure S1 (related to Figure 1): Myo51 and
Gene disruption and construction of C-terminally tagged strains
 Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
The fusion used in most of these studies (MinC4-GFP) contains GFP and
 Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Figure S1. sporulation frequency (%) 80. sme2-5. sme2-3. sme2
 Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
SUPPLEMENTARY INFORMATION
 a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
a 14 12 Densitometry (AU) 1 8 6 4 2 t b 16 NMHC-IIA GAPDH NMHC-IIB Densitometry (AU) 14 12 1 8 6 4 2 1 nm 1 nm 1 nm 1 nm sirna 1 nm 1 nm Figure S1 S4 Quantification of protein levels. (a) The microtubule
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends.
 SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
SUPPLEMENTAL DATA Supplemental Data includes Supplemental Experimental Procedures, 4 Figures and 5 Movie Legends. Supplemental Experimental Procedures Dominant negative nesprin KASH (DN KASH) plasmid construction-
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility
 Current Biology, Volume 25 Supplemental Information RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility Joshua B. Kelley, Gauri Dixit, Joshua B. Sheetz, Sai Phanindra
Current Biology, Volume 25 Supplemental Information RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility Joshua B. Kelley, Gauri Dixit, Joshua B. Sheetz, Sai Phanindra
Supplemental Data. Wang et al. (2017). Plant Cell /tpc NIP1;2 NIP7;1
 Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Supplemental Data. Wang et al. (217). Plant Cell 1.115/tpc.16.825 NIP1;1 NIP1;2 NIP2;1 NIP3;1 NIP4;1 NIP5;1 NIP6;1 NIP7;1 Supplemental Figure 1. Distinct Localization of NIPs in Root Epidermal Cells. (Supports
Orthogonal Ribosome Biofirewall
 1 2 3 4 5 Supporting Information Orthogonal Ribosome Biofirewall Bin Jia,, Hao Qi,, Bing-Zhi Li,, Shuo pan,, Duo Liu,, Hong Liu,, Yizhi Cai, and Ying-Jin Yuan*, 6 7 8 9 10 11 12 Key Laboratory of Systems
1 2 3 4 5 Supporting Information Orthogonal Ribosome Biofirewall Bin Jia,, Hao Qi,, Bing-Zhi Li,, Shuo pan,, Duo Liu,, Hong Liu,, Yizhi Cai, and Ying-Jin Yuan*, 6 7 8 9 10 11 12 Key Laboratory of Systems
Barrales_SuppFig_S1 N/S 5-FOA. no reporter WT N/S 5-FOA. imr::ura4 + lem2. RT-qPCR: subtelomeric genes. lem2. nur1 clr4. WT + empty vector.
 ad f + ad e ad + e6 ch + p cid + + cl r4 + ce ntlh dg / + N/S N/S.5 tell.4.3. clr4 5-FOA + empty vector + empty vector + nmt:lem TZ clr4 RT-qPCR: subtelomeric genes.5.4.3..5. 5 5 4 4 3 3 tell transcript
ad f + ad e ad + e6 ch + p cid + + cl r4 + ce ntlh dg / + N/S N/S.5 tell.4.3. clr4 5-FOA + empty vector + empty vector + nmt:lem TZ clr4 RT-qPCR: subtelomeric genes.5.4.3..5. 5 5 4 4 3 3 tell transcript
Methanol fixation allows better visualization of Kal7. To compare methods for
 Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplementary Data Methanol fixation allows better visualization of Kal7. To compare methods for visualizing Kal7 in dendrites, mature cultures of dissociated hippocampal neurons (DIV21) were fixed with
Supplemental Information. Glycogen Synthesis and Metabolite Overflow. Contribute to Energy Balancing in Cyanobacteria
 Cell Reports, Volume 23 Supplemental Information Glycogen Synthesis and Metabolite Overflow Contribute to Energy Balancing in Cyanobacteria Melissa Cano, Steven C. Holland, Juliana Artier, Rob L. Burnap,
Cell Reports, Volume 23 Supplemental Information Glycogen Synthesis and Metabolite Overflow Contribute to Energy Balancing in Cyanobacteria Melissa Cano, Steven C. Holland, Juliana Artier, Rob L. Burnap,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Kanaani et al., http://www.jcb.org/cgi/content/full/jcb.200912101/dc1 Figure S1. The K2 rabbit polyclonal antibody is specific for GAD67,
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
www.sciencemag.org/cgi/content/full/science.1245533/dc1 Supplementary Materials for A Mechanism for Reorientation of Cortical Microtubule Arrays Driven by Microtubule Severing Jelmer J. Lindeboom, Masayoshi
C. acetobutyricum. Initial co-culture. Transfer 1
 a Co-culture C. acetobutyricum b Initial co-culture Transfer 1 Transfer 2 c Supplementary Figure 1: Persistence of the bacteria in co-culture. (a) Specificity of genes used to follow and in pure culture
a Co-culture C. acetobutyricum b Initial co-culture Transfer 1 Transfer 2 c Supplementary Figure 1: Persistence of the bacteria in co-culture. (a) Specificity of genes used to follow and in pure culture
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
SUPPLEMENTARY INFORMATION
 Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
Nanomechanical mapping of first binding steps of a virus to animal cells David Alsteens, Richard Newton, Rajib Schubert, David Martinez-Martin, Martin Delguste, Botond Roska and Daniel J. Müller This PDF
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 High dynamic range (HDR) imaging of live cells with spinning disk confocal microscopy. During image acquisition, z-sections, encompassing most of the cellular volume, are captured
Supplementary Figure 1 High dynamic range (HDR) imaging of live cells with spinning disk confocal microscopy. During image acquisition, z-sections, encompassing most of the cellular volume, are captured
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
b number of motif occurrences
 supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
supplementary information DOI: 1.138/ncb194 a b number of motif occurrences number of motif occurrences number of motif occurrences I II III IV V X TTGGTCAGTGCA 3 8 23 4 13 239 TTGGTCAGTGCT 7 5 8 1 3 83
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity
 Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
Supplemental figures (1-9) Gu et al. ADF/Cofilin-Mediated Actin Dynamics Regulate AMPA Receptor Trafficking during Synaptic Plasticity Supplemental figure 1. The quantification method to determine the
A legacy of innovation and discovery
 A legacy of innovation and discovery CellInsight CX7 LZR High Content Analysis Platform Quantifiably brilliant data Since the introduction of Thermo Scientific ArrayScan High Content Analysis (HCA) Readers
A legacy of innovation and discovery CellInsight CX7 LZR High Content Analysis Platform Quantifiably brilliant data Since the introduction of Thermo Scientific ArrayScan High Content Analysis (HCA) Readers
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplemental Data. Liu et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Supplemental Figure 1. The GFP Tag Does Not Disturb the Physiological Functions of WDL3. (A) RT-PCR analysis of WDL3 expression in wild-type, WDL3-GFP, and WDL3 (without the GFP tag) transgenic seedlings.
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Supplemental Figure 1 Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical
 Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Supplemental Figure Human REEP family of proteins can be divided into two distinct subfamilies. Residues (single letter amino acid code) identical in all six REEPs are highlighted in green. Additional
Amplicon Sequencing Template Preparation
 Amplicon Sequencing Template Preparation The DNA sample preparation procedure for Amplicon Sequencing consists of a simple PCR amplification reaction, but uses special Fusion Primers (Figure 1-1). The
Amplicon Sequencing Template Preparation The DNA sample preparation procedure for Amplicon Sequencing consists of a simple PCR amplification reaction, but uses special Fusion Primers (Figure 1-1). The
Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by
 Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by deubiquitylases (DUBs) yielding mature Ub deubiquitylase FLAG 3
Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by deubiquitylases (DUBs) yielding mature Ub deubiquitylase FLAG 3
In Situ Hybridization
 In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
Leaf-inspired Artificial Microvascular Networks (LIAMN) for Threedimensional
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 Supporting Information Leaf-inspired Artificial Microvascular Networks (LIAMN) for Threedimensional
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2015 Supporting Information Leaf-inspired Artificial Microvascular Networks (LIAMN) for Threedimensional
Protocols for cell lines using CRISPR/CAS
 Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
Protocols for cell lines using CRISPR/CAS Procedure overview Map Preparation of CRISPR/CAS plasmids Expression vectors for guide RNA (U6-gRNA) and Cas9 gene (CMV-p-Cas9) are ampicillin-resist ant and stable
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
SUPPLEMENTAL INFORMATION: 1. Supplemental methods 2. Supplemental figure legends 3. Supplemental figures
 Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
Supplementary Material (ESI) for Lab on a Chip This journal is The Royal Society of Chemistry 2008 A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Nature Methods: doi: /nmeth.3553
 Supplementary Figure 1 Programming the reconstitution of fully ECM-embedded 3D microtissues by DNA-programmed assembly (DPAC). (a) Timeline for a three-component tissue synthesis by DPAC. The process proceeds
Supplementary Figure 1 Programming the reconstitution of fully ECM-embedded 3D microtissues by DNA-programmed assembly (DPAC). (a) Timeline for a three-component tissue synthesis by DPAC. The process proceeds
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
Supplementary Figure 1. Botrocetin induces binding of human VWF to human
 Supplementary Figure 1: Supplementary Figure 1. Botrocetin induces binding of human VWF to human platelets in the absence of elevated shear and induces platelet agglutination as detected by flow cytometry.
Supplementary Figure 1: Supplementary Figure 1. Botrocetin induces binding of human VWF to human platelets in the absence of elevated shear and induces platelet agglutination as detected by flow cytometry.
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using a spin-column.
 INSTRUCTION MANUAL ZymoPURE Plasmid Miniprep Kit Catalog Nos. D4209, D4210, D4211 & D4212 (Patent Pending) Highlights Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using
INSTRUCTION MANUAL ZymoPURE Plasmid Miniprep Kit Catalog Nos. D4209, D4210, D4211 & D4212 (Patent Pending) Highlights Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 )
 Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Supplementary Figure 1. Espn-1 knockout characterization. (a) The predicted recombinant Espn-1 -/- allele was detected by PCR of the left (5 ) homologous recombination arm (left) and of the right (3 )
Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated
 Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1222901/dc1 Supplementary Materials for Type 6 Secretion Dynamics Within and Between Bacterial Cells M. Basler and J. J. Mekalanos* *To whom correspondence should
www.sciencemag.org/cgi/content/full/science.1222901/dc1 Supplementary Materials for Type 6 Secretion Dynamics Within and Between Bacterial Cells M. Basler and J. J. Mekalanos* *To whom correspondence should
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Fast and efficient site-directed mutagenesis with Platinum SuperFi DNA Polymerase
 APPLICATION NOTE Platinum Superi Polymerase ast and efficient site-directed mutagenesis with Platinum Superi Polymerase Introduction Site-directed mutagenesis is one of the most essential techniques to
APPLICATION NOTE Platinum Superi Polymerase ast and efficient site-directed mutagenesis with Platinum Superi Polymerase Introduction Site-directed mutagenesis is one of the most essential techniques to
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Karotki et al., http://www.jcb.org/cgi/content/full/jcb.201104040/dc1 Figure S1. Eisosome proteins form highly stable filaments of variable
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Karotki et al., http://www.jcb.org/cgi/content/full/jcb.201104040/dc1 Figure S1. Eisosome proteins form highly stable filaments of variable
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive
 Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
Gene specific primers. Left border primer (638) ala3-4 (Salk_082157) Gene specific primers. Left border primer (1343)
 Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Bi 1x Spring 2016: E. coli Growth Curves
 Bi 1x Spring 2016: E. coli Growth Curves 1 Overview In this lab, you will investigate growth of the bacterium E. coli, watching the growth in two ways. First, you will determine growth rate in a solution
Bi 1x Spring 2016: E. coli Growth Curves 1 Overview In this lab, you will investigate growth of the bacterium E. coli, watching the growth in two ways. First, you will determine growth rate in a solution
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Transformation: Theory. Day 2: Transformation Relevant Book Sections
 Day 2: Transformation Relevant Book Sections We will follow the protocols provided in various industry-standard kits, instead of the protocols described in these chapters, but the chapters provide good
Day 2: Transformation Relevant Book Sections We will follow the protocols provided in various industry-standard kits, instead of the protocols described in these chapters, but the chapters provide good
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Strain Genotype Source
 Supplementary Table S1 S. pombe strains used in this study Strain Genotype Source SPDK9 h 90 leu1-32 ura4ds/e ade6-m216 grc3::grc3-egfp-kanmx6 This study SPDK12 h 90 leu1-32 ura4ds/e ade6-m216 grc3::grc3-5flag-kanmx6
Supplementary Table S1 S. pombe strains used in this study Strain Genotype Source SPDK9 h 90 leu1-32 ura4ds/e ade6-m216 grc3::grc3-egfp-kanmx6 This study SPDK12 h 90 leu1-32 ura4ds/e ade6-m216 grc3::grc3-5flag-kanmx6
Franzens-Universitaet Graz, Humboldtstrasse 50, 8010 Graz. Phone: ++43 (0) Fax: ++43 (0)
 Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation Andrea Seper 1, Vera H. I. Fengler 1, Sandro Roier 1, Heimo Wolinski 1, Sepp D. Kohlwein 1, Anne
Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation Andrea Seper 1, Vera H. I. Fengler 1, Sandro Roier 1, Heimo Wolinski 1, Sepp D. Kohlwein 1, Anne
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
Supporting Information. Cationic Conjugated Polymers-Induced Quorum Sensing of Bacteria Cells
 Supporting Information Cationic Conjugated Polymers-Induced Quorum Sensing of Bacteria Cells Pengbo Zhang, Huan Lu, Hui Chen, Jiangyan Zhang, Libing Liu*, Fengting Lv, and Shu Wang* Beijing National Laboratory
Supporting Information Cationic Conjugated Polymers-Induced Quorum Sensing of Bacteria Cells Pengbo Zhang, Huan Lu, Hui Chen, Jiangyan Zhang, Libing Liu*, Fengting Lv, and Shu Wang* Beijing National Laboratory
Supporting Information
 Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs
 Neuron Supplemental Information Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs Frédéric J. Hoerndli, Rui Wang, Jerry E. Mellem, Angy Kallarackal, Penelope J. Brockie,
Neuron Supplemental Information Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs Frédéric J. Hoerndli, Rui Wang, Jerry E. Mellem, Angy Kallarackal, Penelope J. Brockie,
PROTOCOL. Live Cell Imaging of 3D Cultures Grown on Alvetex Scaffold Using Confocal Microscopy. Introduction
 Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
Figure S1 early log mid log late log stat.
 Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Building with DNA 2. Andrew Tolonen Genoscope et l'université d'évry 08 october atolonen at
 Building with DNA 2 Andrew Tolonen Genoscope et l'université d'évry 08 october 2014 atolonen at genoscope.cns.fr @andrew_tolonen www.tolonenlab.org Yesterday we talked about ways to assemble DNA building
Building with DNA 2 Andrew Tolonen Genoscope et l'université d'évry 08 october 2014 atolonen at genoscope.cns.fr @andrew_tolonen www.tolonenlab.org Yesterday we talked about ways to assemble DNA building
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
SUPPLEMENTARY INFORMATION
 Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
File name: Supplementary Information. Description: Supplementary Figures, Supplementary Tables and Supplementary References.
 1 2 File name: Supplementary Information Description: Supplementary Figures, Supplementary Tables and Supplementary References. 3 1 4 Supplementary Figures 5 6 7 Figure S1 Comparison of the One-Step-Assembly
1 2 File name: Supplementary Information Description: Supplementary Figures, Supplementary Tables and Supplementary References. 3 1 4 Supplementary Figures 5 6 7 Figure S1 Comparison of the One-Step-Assembly
Execution of the correct morphogenic program is essential
 FH3, A Domain Found in Formins, Targets the Fission Yeast Formin Fus1 to the Projection Tip During Conjugation Janni Petersen,* Olaf Nielsen,* Richard Egel,* and Iain M. Hagan *Department of Genetics,
FH3, A Domain Found in Formins, Targets the Fission Yeast Formin Fus1 to the Projection Tip During Conjugation Janni Petersen,* Olaf Nielsen,* Richard Egel,* and Iain M. Hagan *Department of Genetics,
indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2% glucose,
 SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells
 Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
Biophysics of contractile ring assembly
 Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
Biophysics of contractile ring assembly Dimitrios Vavylonis Department of Physics, Lehigh University October 1, 2007 Physical biology of the cell Physical processes in cell organization and function: Transport
Supplementary Figure 1
 Supplementary Figure 1 (A) Schematic of sequential hybridization and barcoding. (B) Schematic of the FISH images of the cell. In each round of hybridization, the same spots are detected, but the dye associated
Supplementary Figure 1 (A) Schematic of sequential hybridization and barcoding. (B) Schematic of the FISH images of the cell. In each round of hybridization, the same spots are detected, but the dye associated
In this edition of Marketplace Special offers on Thermo Scientific products:
 In this edition of Marketplace Special offers on Thermo Scientific products: Cloning Enzymes and Kits FastDigest Enzymes GeneJET Nucleic Acid Purification Kits TopVision Agarose Tablets PCR Plastic Consumables
In this edition of Marketplace Special offers on Thermo Scientific products: Cloning Enzymes and Kits FastDigest Enzymes GeneJET Nucleic Acid Purification Kits TopVision Agarose Tablets PCR Plastic Consumables
Complete protocol in 110 minutes Enzymatic fragmentation without sonication One-step fragmentation/tagging to save time
 Molecular Cloning Laboratories Manual Version 1.2 Product name: MCNext UT DNA Sample Prep Kit Cat #: MCUDS-4, MCUDS-24, MCUDS-96 Description: This protocol explains how to prepare up to 96 pooled indexed
Molecular Cloning Laboratories Manual Version 1.2 Product name: MCNext UT DNA Sample Prep Kit Cat #: MCUDS-4, MCUDS-24, MCUDS-96 Description: This protocol explains how to prepare up to 96 pooled indexed
Supplementary methods Shoc2 In Vitro Ubiquitination Assay
 Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary methods Shoc2 In Vitro Ubiquitination Assay 35 S-labelled Shoc2 was prepared using a TNT quick Coupled transcription/ translation System (Promega) as recommended by manufacturer. For the
Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z
 Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z movement and blue piece for x-y movement). Illumination sources
Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z movement and blue piece for x-y movement). Illumination sources
Imaging of BacMam Transfected U-2 OS Cells
 A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
A p p l i c a t i o n N o t e Imaging of BacMam Transfected U-2 OS Cells Optimization of Transfection Conditions Using the Cytation 3 Multi- Mode Reader and Gen5 Data Analysis Software Paul Held Ph. D.
