Figure S1. sporulation frequency (%) 80. sme2-5. sme2-3. sme2
|
|
|
- Jemima Mitchell
- 6 years ago
- Views:
Transcription
1 Figure S1 N WT DSRless mei4 100 ssm4 MS2loop () (kb) sporulation frequency (%) rrna WT DSRless Figure S1. The DSR motifs in are crucial for its function. Expression of the meiotic transcripts was examined by rthern blot analysis in mutants. Wildtype (JS41), -5 (JS42), -3 (JS43) and -DSRless (JS44) cells were grown in MM media at 30 C (N+) and then transferred to MM media without nitrogen source for four hours at 30 C (N-). The rrnas stained with ethidium bromide are shown in the bottom panel as loading controls. Sporulation frequencies of mutants. Wild-type (JS38), (JT926), -5 (JS39), -3' (JS40) and - DSRless (JS5) cells was measured after incubation on an SPA medium at 30 C for 24 hours (n > 200).
2 Figure S2 N WT mei4 mei4 mmi1 pab2 82% (23/28) 18% (5/28) pla1-ts 100% (7/7) rrna clr4 95% (19/20) Figure S2. Localization of in -related mutants. Expression of was examined by rthern blot analysis in mmi1-depleted cells. Wild-type (JY362), mei4 (JZ807) and mmi1 mei4 (JS45) cells were grown in MM media at 30 C (N+) and then transferred to MM media without nitrogen source for four hours at 30 C (N-). The rrnas stained with ethidium bromide are shown in the bottom panel as loading controls. Localization of in mutants involved in -mediated meiotic mrna elimination and facultative heterochromatin formation. pab2 (JS46), pla1-ts (JS47) and clr4 (JS48) cells expressing MS2 loop-tagged from its endogeus promoter and MS2-GFP from the expression vector prep81 were examined under meiotic conditions. Frequencies of meiotic prophase cells containing dot are indicated. Scale bar, 5 µm.
3 Figure S3 1,10- phenanthroline Red1 locus merge colocalization frequency - 94% + 81% 2.0 % input red1 + (untagged) red1-13myc act1 mei Figure S3. Localization of and Red1 at the locus. Localization of and Red1 when transcription is inhibited. Wild-type cells (JS8) expressing Red1-YFP and CFP- from the respective endogeus promoters were grown in YE media and then added with 1,10-phenanthroline (250 µg/ml at final concentration) to inhibit transcription. The LacI-lacO system were used to visualize the locus. Images of the nuclear region after one-hour incubation with or without 1,10-phenanthroline are shown. In the merged image, green indicates Red1-YFP, red indicates the locus, and blue indicates CFP-. Scale bar, 2 µm. Frequencies of cells in which and Red1 localized to the locus are indicated (n > 100). Chromatin immuprecipitation (ChIP) analysis of Red1 accumulation on the act1 locus, the mei4 locus and the 5 or 3 region of the locus in untagged (JY450) and Myc-tagged (JS12) red1 + cells under mitotic conditions. Results represent the mean ± standard deviation from three reactions.
4 Figure S4 locus merge dot formation frequency 1% -5-3 (c) -5 1 dot 2 dots 3 dots 4 dots (%) Figure S4. does t form a dot in mitotic cells without overexpression. Localization of in mitotic cells in which overexpression was t induced. cells (JS16) were incubated in YE medium. The LacI-lacO system were used to visualize the locus. In the merged image, green indicates (MS2-GFP), and red indicates the locus. Scale bar, 5 µm. Frequency of cells containing a dot is indicated (n > 100). Localization of mitotic in cells in which overexpression of variants were t induced. (JS18), -5 (JS19) and -3 (JS20) cells expressing CFP- were incubated in YE media. Scale bar, 5 µm. (c) Percentages of cells containing 1, 2, 3, or 4 and more dots when overexpression of variants were t induced. More than 100 cells were counted in (JS18), -5 (JS19) and -3 (JS20) strains incubated in YE media.
5 Figure S5 + 1 dot 2 dots 3 dots 4 dots + (c) (%) (d) (e) 1 dot 2 dots 3 dots 4 dots (%) (f) -N Figure S5. Mei2 is t involved in the convergence of dots by the overexpression of. Localization of mitotic in cells overexpressing. + (JS21) and (JS22) cells expressing CFP- with the deletion of mei2 were examined under mitotic conditions. Scale bar, 5 µm. Percentages of cells containing 1, 2, 3, or 4 and more dots when variants were overexpressed in cells. More than 100 cells were counted in the + (JS21), (JS22), -5 (JS23), and -3 (JS24) strains with the deletion of mei2 under mitotic conditions. (c) Localization of mitotic in cells overexpressing -5 or (JS23) and -3 (JS24) cells expressing CFP- with the deletion of mei2 were examined under mitotic conditions. Scale bar, 5 µm. (d) Localization of mitotic in cells in which overexpression of variant was t induced. (JS22), -5 (JS23) and -3 (JS24) cells expressing CFP- with the deletion of mei2 were incubated in YE media. Scale bar, 5 µm. (e) Percentages of cells containing 1, 2, 3, or 4 and more dots when overexpression of variants were t induced. More than 100 cells were counted in (JS22), -5 (JS23) and -3 (JS24) strains incubated in YE media. (f) Localization of meiotic in cells overexpressing variants. + (JS21), (JS22), -5 (JS23), and -3 (JS24) cells expressing CFP- with the deletion of mei2 were examined under meiotic conditions. Scale bar, 5 µm.
6 Figure S6 N WT mmi1-ts6 30 C 25 C 28 C 30 C - + mei4 ssm4 rrna rrna Figure S6. The overexpression of reduces the activity of. Expressions of in meiotic wild-type cells and mitotic -overexpressing cells were examined by rthern blot analysis. Wild-type (JY362) cells were grown in MM media at 30 C (N+) and then transferred to MM media without nitrogen source for four hours at 30 C (N-). (JS34) cells were grown in MM media. The rrnas stained with ethidium bromide are shown in the bottom panel as loading controls. Note that overexpressed carries the MS2 loop sequence and is about 130 nt longer than wild-type. Expression of mei4 and ssm4 mrnas was examined by rthern blot analysis in (JS34) and mmi1-ts (JS35) cells with or without overexpression. These cells were grown in YE media (-) and MM media (+) at indicated temperatures. The rrnas stained with ethidium bromide are shown in the bottom panel as loading controls.
7 Table S1. S. pombe strains used in this study Name JS1 JS2 JS3 JS4 JS5 JS6 JS8 JS9 JS10 JS12 JS13 JS14 JS16 JS17 JS18 JS19 JS20 JS21 JS22 JS23 JS24 JS31 JS32 JS33 Getype h 90 lacox32-kan R -ura4 -MS2loop-2 arg1::padh41-4mcherry-laci-nls-hph R CO2::Padh41-MS2-YFP-nat R LEU2-CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 bsd R --MS2loop-2 CO2::Padh41-MS2-YFP-nat R LEU2-CFP-mmi1 mei2-mcherry-hph R ade6-m216 leu1 ura4-d18 h MS2loop-2-Tnmt CO2::Padh41-MS2-YFP-nat R LEU2-CFP-mmi1 mei2-mcherry-hph R ade6-m216 leu1 ura4-d18 h 90 MS2loop-2--3 CO2::Padh41-MS2-YFP-nat R LEU2-CFP-mmi1 mei2-mcherry-hph R ade6-m216 leu1 ura4-d18 h 90 bsd R --DSRless-MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 bsd R --DSRless-MS2loop-2 CO2::Padh41-MS2-YFP-nat R LEU2-CFP-mmi1 mei2-mcherryhph R ade6-m216 leu1 ura4-d18 h 90 lacox32-kan R -ura4 -MS2loop-2 arg1::padh41-4mcherry-laci-nls-hph R LEU2-CFP-mmi1 red1-yfp-nat R ade6-m216 leu1 ura4-d18 h 90 lacox32-kan R -ura4 LEU2--m nat R -CFP-mmi1 red1-mcherry-hph R his7 + <<GFP-lacI ade6-m210 leu1 ura4-d18 h 90 lacox32-kan R -ura4 -DSRless-MS2loop-2 arg1::padh41-4mcherry-laci-nls-hph R LEU2-CFP-mmi1 red1-yfp-nat R ade6-m216 leu1 ura4-d18 h 90 red1-13myc-nat R ade6-m216 leu1 h 90 lacox32-kan R -ura4 LEU2--m red1-13myc-nat R ade6-m216 leu1 ura4-d18 h 90 bsd R --DSRless-MS2loop-2 red1-13myc-nat R ade6-m216 leu1 h 90 lacox32-kan R -ura4 bsd R --MS2loop-2 arg1::padh41-4mcherry-laci-nls-hph R CO2::Padh81-MS2-GFP-nat R ade6-m216 leu1 ura4-d18 h 90 -MS2loop-2-bsd R nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 kan R --MS2loop-2 nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 kan R --5 -MS2loop-2-Tnmt nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 kan R -MS2loop-2--3 nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 mei2::ura4 + nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 mei2::ura4 + kan R --MS2loop-2 nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 mei2::ura4 + kan R --5 -MS2loop-2-Tnmt nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 mei2::ura4 + kan R -MS2loop-2--3 nat R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 CO2::MS2loop-2-ura4-nat R ade6-m216 leu1 ura4-d18 h 90 CO2::MS2loop-2-ssm4-nat R ade6-m216 leu1 ura4-d18 h 90 -MS2loop-2 ade6-m216 leu1 ura4-d18
8 JS34 JS35 JS36 JS37 JS38 JS39 JS40 JS41 JS42 JS43 JS44 JS45 JS46 JS47 JS48 JS50 JS51 JS52 JT926 JV579 JV582 JY362 JY450 JZ807 h 90 kan R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 mmi1-ts6-kan R bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 mei4::ura4 + bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 mei4::ura4 + mmi1::kan R bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 bsd R --5 -MS2loop-2-Tnmt ade6-m216 leu1 ura4-d18 h 90 bsd R --3 -MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 /h 90 bsd R --MS2loop-2/bsd R --MS2loop-2 ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18 h 90 /h 90 bsd R --5 -MS2loop-2-Tnmt/bsd R --5 -MS2loop-2-Tnmt ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18 h 90 /h 90 bsd R --3 -MS2loop-2/bsd R --3 -MS2loop-2 ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18 h 90 /h 90 bsd R --DSRless-MS2loop-2/bsd R --DSRless-MS2loop-2 ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18 h + /h - mei4::ura4 + /mei4::ura4 + mmi1::kan R /mmi1::kan R ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18 h 90 pab2::hph R bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 pla1-41-kan R bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 clr4::kan R bsd R --MS2loop-2 ade6-m216 leu1 ura4-d18 h 90 CO2::MS2loop-2-ura4-nat R kan R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 CO2::MS2loop-2-ssm4-nat R kan R -CFP-mmi1 ade6-m216 leu1 ura4-d18 h 90 mei4-ura4 kan R -lacox32 arg1::padh41-4mcherry-laci-nls-hph R LEU2-CFP-mmi1 ade6-m210 leu1 ura4 h 90 ::ura4 + ade6-m216 leu1 ura4-d18 h 90 mmi1-ts3-kan R ade6-m216 leu1 h 90 mmi1-ts6-kan R ade6-m216 leu1 h - /h + ade6-m216/ade6-m210 leu1/leu1 h 90 ade6-m216 leu1 h - /h + mei4::ura4 + /mei4::ura4 + ade6-m216/ade6-m210 leu1/leu1 ura4-d18/ura4-d18
9 Table S2. Oligonucleotides used in this study For laco integration Name Sequence -laco F1 -laco F2 -laco R2 -laco R1 mei4-laco F1 mei4-laco F2 mei4-laco R2 mei4-laco R1 GTTGCTCCTATAGAAGAACA TTAATTAACCCGGGGATCCGTTCGCGCAGATAAGCGCAAG GTTTAAACGAGCTCGAATTCGATTAAATAGAATATTCGTACATG CACCAAGTTTCTTAAAAGCG ATGAGTCAAGTGGCTACTCAAGTGC TTAATTAACCCGGGGATCCGTTGTTTCGTCAACATATATTATTAC GTTTAAACGAGCTCGAATTCTTCTAAATAAAGTCATAGGAATGTA GCGAGATAGTTAGAAGCGGAAGTGG For quantitative PCR Name Sequence act1-f act1-r mei4-f mei4-r -5 -F -5 -R -3 -F -3 -R TGAGGAGCACCCTTGCTTGT TCTTCTCACGGTTGGATTTGG AATGCGAAACTGAAGCATTG TAGGATCGCCAAACCGATTA AAGACGGAATATGCATGCAAGA AACAAACCACAACACAAAGAAAGAGA GAAAATAAACAATAACCACAGCAAGCT ACAGCACAACCGAAGACCAAT
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast
 Current Biology, Volume 23 Supplemental Information Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast Felipe O. Bendezú and Sophie G. Martin 1 2 Figure S1 (Related to Figure 1). Cells
Current Biology, Volume 23 Supplemental Information Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast Felipe O. Bendezú and Sophie G. Martin 1 2 Figure S1 (Related to Figure 1). Cells
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Supplementary information, Figure S1
 Supplementary information, Figure S1 (A) Schematic diagram of the sgrna and hspcas9 expression cassettes in a single binary vector designed for Agrobacterium-mediated stable transformation of Arabidopsis
Supplementary information, Figure S1 (A) Schematic diagram of the sgrna and hspcas9 expression cassettes in a single binary vector designed for Agrobacterium-mediated stable transformation of Arabidopsis
Caroline Lemieux, Samuel Marguerat, Jennifer Lafontaine, Nicolas Barbezier, Jürg Bähler, and Francois Bachand
 Molecular Cell, Volume 44 Supplemental Information A Pre-mRNA Degradation Pathway that Selectively Targets Intron-Containing Genes Requires the Nuclear Poly(A)-Binding Protein Caroline Lemieux, Samuel
Molecular Cell, Volume 44 Supplemental Information A Pre-mRNA Degradation Pathway that Selectively Targets Intron-Containing Genes Requires the Nuclear Poly(A)-Binding Protein Caroline Lemieux, Samuel
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
7.17: Writing Up Results and Creating Illustrations
 7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive
 Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts
 Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Current Biology, Volume 24 Supplemental Information GM130 Is Required for Compartmental Organization of Dendritic Golgi Outposts Wei Zhou, Jin Chang, Xin Wang, Masha G. Savelieff, Yinyin Zhao, Shanshan
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
THE centromere is a specific chromosomal locus that organizes
 HIGHLIGHTED ARTICLE INVESTIGATION Ectopic Centromere Nucleation by CENP-A in Fission Yeast Marlyn Gonzalez, Haijin He, Qianhua Dong, Siyu Sun, and Fei Li 1 Department of Biology, New York University, New
HIGHLIGHTED ARTICLE INVESTIGATION Ectopic Centromere Nucleation by CENP-A in Fission Yeast Marlyn Gonzalez, Haijin He, Qianhua Dong, Siyu Sun, and Fei Li 1 Department of Biology, New York University, New
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli
 Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplemental Data. Regulating Gene Expression. through RNA Nuclear Retention
 Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of
 Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Supplemental Information. Boundary Formation through a Direct. Threshold-Based Readout. of Mobile Small RNA Gradients
 Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Developmental Cell, Volume 43 Supplemental Information Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients Damianos S. Skopelitis, Anna H. Benkovics, Aman Y. Husbands,
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supplemental Material for Xue et al. List. Supplemental Figure legends. Figure S1. Related to Figure 1. Figure S2. Related to Figure 3
 Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
SUPPLEMENTARY INFORMATION FOR SELA ET AL., Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans
 SUPPLEMENTARY INFORMATION FOR SELA ET AL., Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans Supplementary methods Plasmids and strains construction CaRUB1
SUPPLEMENTARY INFORMATION FOR SELA ET AL., Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans Supplementary methods Plasmids and strains construction CaRUB1
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
 Watson BM Gene Capitolo 11 Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. ? Le proteine della trasposizione Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A.
Watson BM Gene Capitolo 11 Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. ? Le proteine della trasposizione Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A.
7.06 Problem Set #3, Spring 2005
 7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
7.06 Problem Set #3, Spring 2005 1. The Drosophila compound eye is composed of about 800 units called ommatidia. Each ommatidium contains eight photoreceptor neurons (R1 through R8), which develop in a
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Active-Site Mutations in the Xrn1p Exoribonuclease of Saccharomyces cerevisiae Reveal a Specific Role in Meiosis
 MOLECULAR AND CELLULAR BIOLOGY, Sept. 1999, p. 5930 5942 Vol. 19, No. 9 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Active-Site Mutations in the Xrn1p
MOLECULAR AND CELLULAR BIOLOGY, Sept. 1999, p. 5930 5942 Vol. 19, No. 9 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Active-Site Mutations in the Xrn1p
Stable mrnp Formation and Export Require Cotranscriptional Recruitment of the mrna Export Factors Yra1p and Sub2p by Hpr1p
 MOLECULAR AND CELLULAR BIOLOGY, Dec. 2002, p. 8241 8253 Vol. 22, No. 23 0270-7306/02/$04.00 0 DOI: 10.1128/MCB.22.23.8241 8253.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, Dec. 2002, p. 8241 8253 Vol. 22, No. 23 0270-7306/02/$04.00 0 DOI: 10.1128/MCB.22.23.8241 8253.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved.
Analysis of two mitochondrial functions in Saccharomyces cerevisiae
 PhD thesis Analysis of two mitochondrial functions in Saccharomyces cerevisiae Zsuzsanna Fekete Supervisors: Gyula Kispál, MD, PhD, Katalin Sipos, MD, PhD University of Pécs Medical School Pécs, 2010 Introduction
PhD thesis Analysis of two mitochondrial functions in Saccharomyces cerevisiae Zsuzsanna Fekete Supervisors: Gyula Kispál, MD, PhD, Katalin Sipos, MD, PhD University of Pécs Medical School Pécs, 2010 Introduction
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
 Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
 Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes Isabelle Toesca., Camille R. Nery., Cesar F. Fernandez, Shakir Sayani, Guillaume F. Chanfreau* Department of Chemistry
Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes Isabelle Toesca., Camille R. Nery., Cesar F. Fernandez, Shakir Sayani, Guillaume F. Chanfreau* Department of Chemistry
Transforming Growth Factor -Independent Shuttling of Smad4 between the Cytoplasm and Nucleus
 MOLECULAR AND CELLULAR BIOLOGY, Dec. 2000, p. 9041 9054 Vol. 20, No. 23 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Transforming Growth Factor -Independent
MOLECULAR AND CELLULAR BIOLOGY, Dec. 2000, p. 9041 9054 Vol. 20, No. 23 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Transforming Growth Factor -Independent
H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases
 H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases Klavs R. Hansen 1,2, Idit Hazan 3, Sreenath Shanker 4, Stephen Watt 5,6, Janne Verhein-Hansen 1,Jürg
H3K9me-Independent Gene Silencing in Fission Yeast Heterochromatin by Clr5 and Histone Deacetylases Klavs R. Hansen 1,2, Idit Hazan 3, Sreenath Shanker 4, Stephen Watt 5,6, Janne Verhein-Hansen 1,Jürg
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Prions Other molecules besides organelle DNA are inherited in non-mendelian patterns.
 Prions Other molecules besides organelle DNA are inherited in non-mendelian patterns. Examples of non-mendelian patterns of inheritance extend beyond the inheritance of organelle DNA. Certain DNA and RNA
Prions Other molecules besides organelle DNA are inherited in non-mendelian patterns. Examples of non-mendelian patterns of inheritance extend beyond the inheritance of organelle DNA. Certain DNA and RNA
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility
 Current Biology, Volume 25 Supplemental Information RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility Joshua B. Kelley, Gauri Dixit, Joshua B. Sheetz, Sai Phanindra
Current Biology, Volume 25 Supplemental Information RGS Proteins and Septins Cooperate to Promote Chemotropism by Regulating Polar Cap Mobility Joshua B. Kelley, Gauri Dixit, Joshua B. Sheetz, Sai Phanindra
Supplemental Information. Antagonistic Activities of Sox2. and Brachyury Control the Fate Choice. of Neuro-Mesodermal Progenitors
 Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
Developmental Cell, Volume 42 Supplemental Information Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors Frederic Koch, Manuela Scholze, Lars Wittler,
Supporting Information
 Supporting Information Rougemaille et al. 1.173/pnas.12494719 SI Methods Strain Construction. Homologous replacement of DNA was accomplished by lithium acetate transformation of PCR products containing
Supporting Information Rougemaille et al. 1.173/pnas.12494719 SI Methods Strain Construction. Homologous replacement of DNA was accomplished by lithium acetate transformation of PCR products containing
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis
 DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis Sizolwenkosi Mlotshwa 1, Gail J. Pruss 1, Angela Peragine 2, Matthew W. Endres 1, Junjie Li 3, Xuemei Chen 3, R. Scott
DICER-LIKE2 Plays a Primary Role in Transitive Silencing of Transgenes in Arabidopsis Sizolwenkosi Mlotshwa 1, Gail J. Pruss 1, Angela Peragine 2, Matthew W. Endres 1, Junjie Li 3, Xuemei Chen 3, R. Scott
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
TECHNICAL BULLETIN. U2OS GFP-NUP98 Osteosarcoma Cell Line with 3XFLAG -GFPtagged. Catalog Number CLL1136 Storage Temperature 196 C (liquid nitrogen)
 U2OS GFP-NUP98 Osteosarcoma Cell Line with 3XFLAG -GFPtagged NUP98 Catalog Number CLL1136 Storage Temperature 196 C (liquid nitrogen) TECHNICAL BULLETIN Product Description This product is a human U2OS
U2OS GFP-NUP98 Osteosarcoma Cell Line with 3XFLAG -GFPtagged NUP98 Catalog Number CLL1136 Storage Temperature 196 C (liquid nitrogen) TECHNICAL BULLETIN Product Description This product is a human U2OS
TTTCAGGGTCCATAAGATCCTAGGGATAACAGGGTAATAGATCTAAGCTTTTC AATTCA
 Supplementary data Materials and methods Strains and plasmids The strains used in this study are listed in supplementary Table 1. Gene deletions were made by PCR-mediated transformation with the pfa6a-skhis3-mx6,
Supplementary data Materials and methods Strains and plasmids The strains used in this study are listed in supplementary Table 1. Gene deletions were made by PCR-mediated transformation with the pfa6a-skhis3-mx6,
Keiko Ogino and Hisao Masai 1 From the Genome Dynamics Project, Tokyo Metropolitan Institute of Medical Science, Tokyo , Japan
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 281, NO. 3, pp. 1338 1344, January 20, 2006 2006 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Rad3-Cds1 Mediates Coupling
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 281, NO. 3, pp. 1338 1344, January 20, 2006 2006 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Rad3-Cds1 Mediates Coupling
Universitaet Regensburg, Institut für Biochemie, Genetik und Mikrobiologie, Regensburg, Germany
 Actively transcribed rrna genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules Katharina Merz,
Actively transcribed rrna genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules Katharina Merz,
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Sordaria Lab Report. Sordaria Fimicola reproduce through the process of meiosis. This process entails
 Adil Sabir Bio 110H 18 October 2014 Sordaria Lab Report I. Introduction Sordaria Fimicola reproduce through the process of meiosis. This process entails prophase I, metaphase I, anaphase I, telophase I,
Adil Sabir Bio 110H 18 October 2014 Sordaria Lab Report I. Introduction Sordaria Fimicola reproduce through the process of meiosis. This process entails prophase I, metaphase I, anaphase I, telophase I,
Supporting Information
 Supporting Information Beauregard et al. 10.1073/pnas.1218984110 Fig. S1. Model depicting the regulatory pathway leading to expression of matrix genes. The master regulator Spo0A can be directly or indirectly
Supporting Information Beauregard et al. 10.1073/pnas.1218984110 Fig. S1. Model depicting the regulatory pathway leading to expression of matrix genes. The master regulator Spo0A can be directly or indirectly
Inhibition of TATA-Binding Protein Function by SAGA Subunits Spt3 and Spt8 at Gcn4-Activated Promoters
 MOLECULAR AND CELLULAR BIOLOGY, Jan. 2000, p. 634 647 Vol. 20, No. 2 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Inhibition of TATA-Binding Protein Function
MOLECULAR AND CELLULAR BIOLOGY, Jan. 2000, p. 634 647 Vol. 20, No. 2 0270-7306/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Inhibition of TATA-Binding Protein Function
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
No, because expression of the P elements and hence transposase in suppressed in the F1.
 Problem set B 1. A wild-type ry+ (rosy) gene was introduced into a ry mutant using P element-mediated gene transformation, and a strain containing a stable ry+ gene was established. If a transformed male
Problem set B 1. A wild-type ry+ (rosy) gene was introduced into a ry mutant using P element-mediated gene transformation, and a strain containing a stable ry+ gene was established. If a transformed male
Product Name : Simple mirna Detection Kit
 Product Name : Simple mirna Detection Kit Code No. : DS700 This product is for research use only Kit Contents This kit provides sufficient reagents to perform 20 reactions for detecting microrna. Components
Product Name : Simple mirna Detection Kit Code No. : DS700 This product is for research use only Kit Contents This kit provides sufficient reagents to perform 20 reactions for detecting microrna. Components
B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast
 B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast Mark K. Chee, Steven B. Haase* Department of Biology, Duke University, Durham, North Carolina, United States
B-Cyclin/CDKs Regulate Mitotic Spindle Assembly by Phosphorylating Kinesins-5 in Budding Yeast Mark K. Chee, Steven B. Haase* Department of Biology, Duke University, Durham, North Carolina, United States
Fig. S1. Nature Medicine: doi: /nm HoxA9 expression levels BM MOZ-TIF2 AML BM. Sca-1-H. c-kit-h CSF1R-H CD16/32-H. Mac1-H.
 A 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 1 1 1 1 1 2 1 3 1 4 GFPH 1 1 1 1 1 2 1 3 1 4 Sca1H 1 1 1 1 1 2 1 3 1 4 ckith 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH
A 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 1 1 1 1 1 2 1 3 1 4 GFPH 1 1 1 1 1 2 1 3 1 4 Sca1H 1 1 1 1 1 2 1 3 1 4 ckith 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley
 Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
Targeted modification of gene function exploiting homology directed repair of TALENmediated double strand breaks in barley Nagaveni Budhagatapalli a, Twan Rutten b, Maia Gurushidze a, Jochen Kumlehn a,
1A) What is different about the resulting resection of ends by these two complexes?
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. Your lab is studying the MRX complex in S. pombe, otherwise known as fission yeast. For your dissertation work, you are studying
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. Your lab is studying the MRX complex in S. pombe, otherwise known as fission yeast. For your dissertation work, you are studying
Green Fluorescent Protein. Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani
 Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Tension directly stabilizes reconstituted. kinetochore-microtubule attachments
 SUPPLEMENTARY INFORMATION Tension directly stabilizes reconstituted kinetochore-microtubule attachments Bungo Akiyoshi, Krishna K. Sarangapani, Andrew F. Powers, Christian R. Nelson, Steve L. Reichow,
SUPPLEMENTARY INFORMATION Tension directly stabilizes reconstituted kinetochore-microtubule attachments Bungo Akiyoshi, Krishna K. Sarangapani, Andrew F. Powers, Christian R. Nelson, Steve L. Reichow,
Supporting Information. SI Material and Methods
 Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the levels of Exo1
 6530 6545 Nucleic Acids Research, 2017, Vol. 45, No. 11 Published online 2 May 2017 doi: 10.1093/nar/gkx347 The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the
6530 6545 Nucleic Acids Research, 2017, Vol. 45, No. 11 Published online 2 May 2017 doi: 10.1093/nar/gkx347 The RNA binding protein Npl3 promotes resection of DNA double-strand breaks by regulating the
Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila
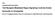 Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
Cell Supplemental Information Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila Bo Liu, Yonggang Zheng, Feng Yin, Jianzhong Yu, Neal Silverman, and Duojia Pan Supplemental Experimental
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis
 Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
Impact of Retinoic acid induced-1 (Rai1) on Regulators of Metabolism and Adipogenesis The mammalian system undergoes ~24 hour cycles known as circadian rhythms that temporally orchestrate metabolism, behavior,
A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
 Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Immunofluorescence images of different core histones and different histone exchange assay.
 Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
Molecular Cell, Volume 51 Supplemental Information Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage Christoffel Dinant, Giannis Ampatziadis-Michailidis,
The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control
 The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control Alan J. Bridge, Mary Morphew, 1 Rachel Bartlett, 2 and Iain M. Hagan 3 School of Biological Sciences, University
The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control Alan J. Bridge, Mary Morphew, 1 Rachel Bartlett, 2 and Iain M. Hagan 3 School of Biological Sciences, University
Supplement Figure 1. Characterization of the moab. (A) A series of moabs that are anti-hαiib-specific were tested for their ability to bind to
 Supplement Figure 1. Characterization of the 312.8 moab. (A) A series of moabs that are anti-hαiib-specific were tested for their ability to bind to platelets. The black line represents the 312.8 moab
Supplement Figure 1. Characterization of the 312.8 moab. (A) A series of moabs that are anti-hαiib-specific were tested for their ability to bind to platelets. The black line represents the 312.8 moab
R1 12 kb R1 4 kb R1. R1 10 kb R1 2 kb R1 4 kb R1
 Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
Bcor101 Sample questions Midterm 3 1. The maps of the sites for restriction enzyme EcoR1 (R1) in the wild type and mutated cystic fibrosis genes are shown below: Wild Type R1 12 kb R1 4 kb R1 _ _ CF probe
6.2 Chromatin is divided into euchromatin and heterochromatin
 6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
6.2 Chromatin is divided into euchromatin and heterochromatin Individual chromosomes can be seen only during mitosis. During interphase, the general mass of chromatin is in the form of euchromatin. Euchromatin
Supplementary Figure 1. Comparison of nodules from Gifu and epr3-9 plants inoculated with M. loti MAFF and incubated at 21 C or 28 C.
 Supplementary Figure 1. Comparison of nodules from Gifu and epr3-9 plants inoculated with M. loti MAFF303099 and incubated at 21 C or 28 C. (a to l) are at 21 C, and (m to å) are at 28 C. (a, e, i, m,
Supplementary Figure 1. Comparison of nodules from Gifu and epr3-9 plants inoculated with M. loti MAFF303099 and incubated at 21 C or 28 C. (a to l) are at 21 C, and (m to å) are at 28 C. (a, e, i, m,
Confirming the Phenotypes of E. coli Strains
 Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Analysis of TFIIA Function In Vivo: Evidence for a Role in TATA-Binding Protein Recruitment and Gene-Specific Activation
 MOLECULAR AND CELLULAR BIOLOGY, Dec. 1999, p. 8673 8685 Vol. 19, No. 12 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Analysis of TFIIA Function In Vivo:
MOLECULAR AND CELLULAR BIOLOGY, Dec. 1999, p. 8673 8685 Vol. 19, No. 12 0270-7306/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Analysis of TFIIA Function In Vivo:
Supplemental Fig. S1. Key to underlines: Key to amino acids:
 AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
AspA-F1 AspA 1 MKQMETKGYGYFRKTKAYGLVCGIT--------------LAGALTLGTTSVSADDVTTLNPATNLTTLQTPPTADQTQLAHQAGQQSGELVSEVSNTEWD 86 SspB 1 MQKREV--FG-FRKSKVAKTLCGAV-LGAALIAIADQQVLADEVTETNSTANVAVTTTGNPATNLPEAQGEATEAASQSQAQAGSKDGALPVEVSADDLN
TECHNICAL BULLETIN. GenElute mrna Miniprep Kit. Catalog MRN 10 MRN 70
 GenElute mrna Miniprep Kit Catalog Numbers MRN 10, MRN 70 TECHNICAL BULLETIN Product Description The GenElute mrna Miniprep Kit provides a simple and convenient way to purify polyadenylated mrna from previously
GenElute mrna Miniprep Kit Catalog Numbers MRN 10, MRN 70 TECHNICAL BULLETIN Product Description The GenElute mrna Miniprep Kit provides a simple and convenient way to purify polyadenylated mrna from previously
The mitotic spindle is required for loading of the DASH complex onto the kinetochore
 The mitotic spindle is required for loading of the DASH complex onto the kinetochore Yumei Li, 1 Jeff Bachant, 1 Annette A.Alcasabas, 1 Yanchang Wang, 1 Jun Qin, 1,2 and Stephen J.Elledge 1,3,4,5 1 Verna
The mitotic spindle is required for loading of the DASH complex onto the kinetochore Yumei Li, 1 Jeff Bachant, 1 Annette A.Alcasabas, 1 Yanchang Wang, 1 Jun Qin, 1,2 and Stephen J.Elledge 1,3,4,5 1 Verna
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Methods. Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Supplementary Methods Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated by introducing a linker containing an XbaI and
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Supplementary Methods Plasmid construction. NS5A-fluorophore fusion Jc1 genomes were generated by introducing a linker containing an XbaI and
Supplemental Data. Distinct Pathways for snorna and mrna Termination
 Molecular Cell, Volume 24 Supplemental Data Distinct Pathways for snorna and mrna Termination Minkyu Kim, Lidia Vasiljeva, Oliver J. Rando, Alexander Zhelkovsky, Claire Moore, and Stephen Buratowski A
Molecular Cell, Volume 24 Supplemental Data Distinct Pathways for snorna and mrna Termination Minkyu Kim, Lidia Vasiljeva, Oliver J. Rando, Alexander Zhelkovsky, Claire Moore, and Stephen Buratowski A
Summary of Mutagenic Toxicity Test Results for EvaGreen
 Summary of Mutagenic Toxicity Test Results for EvaGreen Compiled by Biotium, Inc. from the results of an independent testing service: Litron Laboratories, Inc., Rochester, NY Overview When our scientists
Summary of Mutagenic Toxicity Test Results for EvaGreen Compiled by Biotium, Inc. from the results of an independent testing service: Litron Laboratories, Inc., Rochester, NY Overview When our scientists
Endoplasmic Reticulum Stress Induction of the Grp78/BiP Promoter: Activating Mechanisms Mediated by YY1 and Its Interactive Chromatin Modifiers
 MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4529 4540 Vol. 25, No. 11 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.11.4529 4540.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, June 2005, p. 4529 4540 Vol. 25, No. 11 0270-7306/05/$08.00 0 doi:10.1128/mcb.25.11.4529 4540.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved.
RITS acts in cis to promote RNA interference mediated transcriptional and post-transcriptional silencing
 00 Nature Publishing Group http://www.nature.com/naturegenetics RITS acts in cis to promote RNA interference mediated transcriptional and post-transcriptional silencing Ken-ichi Noma, Tomoyasu Sugiyama,
00 Nature Publishing Group http://www.nature.com/naturegenetics RITS acts in cis to promote RNA interference mediated transcriptional and post-transcriptional silencing Ken-ichi Noma, Tomoyasu Sugiyama,
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Transcriptional Activation of the General Amino Acid Permease Gene per1 by the Histone Deacetylase Clr6 Is Regulated by Oca2 Kinase
 MOLECULAR AND CELLULAR BIOLOGY, July 2010, p. 3396 3410 Vol. 30, No. 13 0270-7306/10/$12.00 doi:10.1128/mcb.00971-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Transcriptional
MOLECULAR AND CELLULAR BIOLOGY, July 2010, p. 3396 3410 Vol. 30, No. 13 0270-7306/10/$12.00 doi:10.1128/mcb.00971-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Transcriptional
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Center Drive, University of Michigan Health System, Ann Arbor, MI
 Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
 Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1 Yves B. Beaulieu 1, Claudia L. Kleinman 2, Anne-Marie Landry-Voyer 1, Jacek Majewski 2, François
Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1 Yves B. Beaulieu 1, Claudia L. Kleinman 2, Anne-Marie Landry-Voyer 1, Jacek Majewski 2, François
Problem Set 2B Name and Lab Section:
 Problem Set 2B 9-26-06 Name and Lab Section: 1. Define each of the following rearrangements (mutations) (use one phrase or sentence for each). Then describe what kind of chromosomal structure you might
Problem Set 2B 9-26-06 Name and Lab Section: 1. Define each of the following rearrangements (mutations) (use one phrase or sentence for each). Then describe what kind of chromosomal structure you might
DNA Structure and Properties Basic Properties Predicting Melting Temperature. Dinesh Yadav
 DNA Structure and Properties Basic Properties Predicting Melting Temperature Dinesh Yadav Nucleic Acid Structure Question: Is this RNA or DNA? Molecules of Life, pp. 15 2 Nucleic Acid Bases Molecules of
DNA Structure and Properties Basic Properties Predicting Melting Temperature Dinesh Yadav Nucleic Acid Structure Question: Is this RNA or DNA? Molecules of Life, pp. 15 2 Nucleic Acid Bases Molecules of
Mechanism of Prion Loss after Hsp104 Inactivation in Yeast
 MOLECULAR AND CELLULAR BIOLOGY, July 2001, p. 4656 4669 Vol. 21, No. 14 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.14.4656 4669.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, July 2001, p. 4656 4669 Vol. 21, No. 14 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.14.4656 4669.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
