Supplementary Materials: Viral Protein Kinetics of Piscine Orthoreovirus Infection in Atlantic Salmon Blood Cells
|
|
|
- Paulina Lambert
- 6 years ago
- Views:
Transcription
1 S1of S7 Supplementary Materials: Viral Protein Kinetics of Piscine Orthoreovirus Infection in Atlantic Salmon Blood Cells Hanne Merethe Haatveit, Øystein Wessel, Turhan Markussen, Morten Lund, Bernd Thiede, Ingvild Berg Nyman, Stine Braaen, Maria Krudtaa Dahle and Espen Rimstad Figure S1. Specificity of μns and λ1 antisera. (a) The specificity of the μns and λ1 antisera were verified by western blotting of lysates from EPC cells transfected with plasmids encoding μns or λ1, or mock transfected. Anti rabbit IgG HRP conjugate was used as secondary antibody. M = molecular weight standard. The theoretical MWs of μns and λ1 are 83.5 and kda, respectively. (b) Staining of μns (left) and λ1 (right) in transfected EPC cells using rabbit IgG conjugated with Alexa Fluor 488 (green) as secondary antibody. The nuclei were stained with Hoechst (blue). Figure S2. PRV RNA load in blood cells (second challenge experiment). RT qpcr of PRV gene segment S1 in blood cells from cohabitant fish. Individual (dots) and mean (line) Ct values, n = 6 per time point. wpc = weeks post challenge.
2 S2of S7 Figure S3. Expression of immune genes in blood cells. Immune genes were assayed at 3 8 wpc by RTqPCR in blood cells from cohabitant fish (n= 6 per time point). Data are normalized against EF1α and the mean ΔCt level of each target at 3 wpc (n=6), and 2 ΔΔCt values are calculated. Relative expression +/ SD is shown. wpc = weeks post challenge, IFN = interferon. 30 EF1a WPC Figure S4. Expression of EF1α during PRV infection.
3 S3of S7 Figure S5. PRV σ1 positive blood cells detected by flow cytometry. Flow cytometry result showing intracellular staining of σ1 in blood cells from three cohabitant fish sampled 4, 5 and 6 wpc. One fish sampled 0 wpc is presented as negative control cells were counted for each sample and cells were gated for analysis.
4 S4of S7 Figure S6. Presence of PRV proteins in blood cells (second challenge experiment). Pooled blood cell samples (n = 6) from each week were analyzed by western blotting targeting μns, σ1, σ3, μ1 and λ1. M = molecular weight standard, wpc = weeks post challenge.
5 S5of S7 Figure S7. LC MS analyses of PRV μns. (a) PRV μns peptide sequences identified following IP with anti μns antisera, SDS PAGE, excision of the 70 kda (4 wpc) and 83.5 kda (5 wpc) fragments, tryptic digestion and LC MS analysis. Peptides identified from the 83.5 kda fragment are shown with blue background, while peptides derived from both fragments are with green background. Peptides exclusive to the 70 kda band were not identified. Numbers in italic indicate peptide spectrum matches derived from both bands, i.e. in the order 83.5 kda/70kda. The putative secondary in frame translation initiation site encoding M115 is shown in bold and underlined. (b) PRV μns peptide spectrum matches identified in the 200 aa N and the 552 aa C terminal regions of μns, derived from the 70 kda and 83.5 kda bands.
6 S6of S7 Figure S8. LC MS analyses of PRV μ1. (a) PRV μ1 peptide sequences identified following IP with antiμ1c antisera 5 wpc, SDS PAGE, excision of the 70 kda, 37 kda and 32 kda fragments, tryptic digestion and LC MS analysis, shown with orange, blue and red backgrounds, respectively. Green background indicate peptides identified from all three fragments (common peptide sequences from two bands only were not observed). The cleavage site N42P43 and the putative cleavage site at F387S388 are shown in bold and underlined. Numbers in italic indicate peptide spectrum matches derived from the three bands, i.e. in the order 70kDa/37kDa/32kDa. *Non tryptic N terminal cleavage site. **Non tryptic C terminal cleavage site. (b) PRV μ1 peptide spectrum matches N and C terminal to S388F387 identified from the 70, 37 and 32 kda bands.
7 S7of S7 Table S1. Primers and probes used for construction of plasmids and expression of viral RNA levels. Plasmid/target Primer sequence (5 3 ) pentr μns Fwd CACCATGGCTGAATCAATTACTTTTGGAGGA Rev GCCACGTAGCACATTATTCACGCCCAC pet100 λ1 Fwd CACCATGGAGCGACTTAAGAGGAA Rev TTAGTTGAGTACAGGATGAGTCAA Fwd: GCGTCCTGCGTATGGCACC Rev: GGCTGGCAT GCCCGAATAGCA S1 Probe ATCACAACGCCTACCT MGBNFQ (FAM): Fwd: CCCTCAAGCCAACCAATGTGTGCTA Rev: GTGTCAACATTCCTGACAGTCGGTGAA M2 Probe TAGCTCGAATCACTTGCAG MGBNFQ (FAM): Fwd: GTCATCTGGAGTCGTTCTGCCACT Rev: GCACACTGCCAGGAATCCCACT M3 Probe TTCATCACGTCTTGCG MGBNFQ (FAM): Table S2. Primers used for analysis of antiviral gene expression. Target gene EF1α IFNab Mx Viperin ISG15 sspkr IFNγ CD8α CD4 Primer/probe sequence (5 3 ) Fwd: TGCCCCTCCAGGATGTCTAC Rev: CACGGCCCACAGGTACTG Fwd: CCTTTCCCTGCTGGACCA Rev: TGTCTGTAAAGGGATGTTGGGAAAA Fwd: GATGCTGCACCTCAAGTCCTATTA Rev: CACCAGGTAGCGGATCACCAT Fwd: AGCAATGGCAGCATGATCAG Rev: TGGTTGGTGTCCTCGTCAAAG Fwd: GGCCTGCATTCAGGATCTAA Rev: TACAGTCTCACCAGGCACCA Fwd: CAGGATGCAACACCATCATC Rev: GGTCTTGACCGGTGACATCT Fwd: AAGGGCTGTGATGTGTTTCTG Rev: TGTACTGAGCGGCATTACTCC Fwd: GTCTACAGCTGTGCATCAATCAA Rev: GGCTGTGGTCATTGGTGTAGT Fwd GCCCCTGAAGTCCAACGAC Rev: AGGCTTCTCTCACTGCGTCC Genbank acc. no. BG DQ XM_ NM_ AY EF NM_ NM_ NM_ by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC BY) license (
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells.
 Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
This is the author's accepted version of the manuscript.
 This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
This is the author's accepted version of the manuscript. The definitive version is published in Nature Communications Online Edition: 2015/4/16 (Japan time), doi:10.1038/ncomms7780. The final version published
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Viral RNAi suppressor reversibly binds sirna to. outcompete Dicer and RISC via multiple-turnover
 Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated
 A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells
 SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Product Data Sheet - ANTIBODY
 888.267.4436 techsupport@origene.com www.origene.com Name:Mouse Monoclonal MBP Antibody (2H9) Product Data Sheet - ANTIBODY Catalog: TA336919 Components: Mouse Monoclonal MBP Antibody (2H9) (TA336919)
888.267.4436 techsupport@origene.com www.origene.com Name:Mouse Monoclonal MBP Antibody (2H9) Product Data Sheet - ANTIBODY Catalog: TA336919 Components: Mouse Monoclonal MBP Antibody (2H9) (TA336919)
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Western-GUARANTEED Antibody Service FAQ
 Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A
 Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A schematic of the eye showing the ciliary margin (CM) of the anterior region, and the neuroepithelium (NE) and retinal
Supplementary Figure S1. Expression of complement components in E4 chick eyes. (a) A schematic of the eye showing the ciliary margin (CM) of the anterior region, and the neuroepithelium (NE) and retinal
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure 1 a
 3 min PMA 45 min PMA AnnexinV-FITC Supplementary Figure 1 5 min PMA 15 min PMA a 9 min PMA 12 min PMA 5 min FGF7 15 min FGF7 3 min FGF7 6 min FGF7 9 min FGF7 12 min FGF7 5 min control 3 min control 6 min
3 min PMA 45 min PMA AnnexinV-FITC Supplementary Figure 1 5 min PMA 15 min PMA a 9 min PMA 12 min PMA 5 min FGF7 15 min FGF7 3 min FGF7 6 min FGF7 9 min FGF7 12 min FGF7 5 min control 3 min control 6 min
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Cell viability. Cell viability was examined by MTT assay (Sigma-Aldrich).
 Supplementary Materials Supplementary materials and methods Cell culture. Primary human dermal fibroblasts (DFs) were isolated from full-thickness skin samples. Tissue samples were dissected into small
Supplementary Materials Supplementary materials and methods Cell culture. Primary human dermal fibroblasts (DFs) were isolated from full-thickness skin samples. Tissue samples were dissected into small
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure
 ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplemental Figure 1 Kam et al., 2017
 nti-zikv IgG response (OD 450 nm) % Infectivity (Relative to virus only) 3 2 1 2 0 1 0 0 8 0 6 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C-LL 1 4 0 2 0 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C -LL 0 0.1 1 1 0 1 0 0 [b],
nti-zikv IgG response (OD 450 nm) % Infectivity (Relative to virus only) 3 2 1 2 0 1 0 0 8 0 6 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C-LL 1 4 0 2 0 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C -LL 0 0.1 1 1 0 1 0 0 [b],
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTARY INFORMATION
 ARTICLE NUMBER: 0 DOI: 0.0/NMICROBIOL.0. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes Tomomi Ando,, Seiya Yamayoshi, Yuriko Tomita,, Shinji
ARTICLE NUMBER: 0 DOI: 0.0/NMICROBIOL.0. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes Tomomi Ando,, Seiya Yamayoshi, Yuriko Tomita,, Shinji
Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein
 Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein Maybelline Giam 1,2, Toru Okamoto 1,2,3, Justine D. Mintern 1,2,4, Andreas Strasser 1,2 and Philippe Bouillet 1, 2 1 The Walter and Eliza
Bcl-2 family member Bcl-G is not a pro-apoptotic BH3-only protein Maybelline Giam 1,2, Toru Okamoto 1,2,3, Justine D. Mintern 1,2,4, Andreas Strasser 1,2 and Philippe Bouillet 1, 2 1 The Walter and Eliza
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence
 Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Supplementary Figure Legends Supplementary Fig S1 Nutlin-3a treatment does not affect cell cycle progression in the absence of p53 or p21. HCT116 cells which were null for either p53 (A) or p21 (B) were
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Gene Forward Primer Reverse Primer GAPDH ATCATCCCTGCCTCTACTGG GTCAGGTCCACCACTGACAC SSB1 AACTTCAGTGAGCCAAACCC GTTCTCAGAGGCTGGAGAGG
 Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
Supplemental Data EXPERIMENTAL PROCEDURES Plasmids and Antibodies- Full length cdna of INT11 or INT12 were cloned into ps- Flag-SBP vector respectively. Anti-RNA pol II (RPB1) was purchased from Santa
m/z (mass to charge ratio)
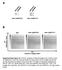 a Anti-GANP(N1) Anti-GANP(C1) b Anti-GANP(N1) Anti-GANP(C1) RT (Retention Time) IgG m/z (mass to charge ratio) Supplementary Figure S1. 2DICAL analysis of GANP complex from Ramos nuclei. (a) Specificity
a Anti-GANP(N1) Anti-GANP(C1) b Anti-GANP(N1) Anti-GANP(C1) RT (Retention Time) IgG m/z (mass to charge ratio) Supplementary Figure S1. 2DICAL analysis of GANP complex from Ramos nuclei. (a) Specificity
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Agglutinating mouse IgG3 compares favourably with IgMs in typing of the blood group B
 Agglutinating mouse IgG3 compares favourably with IgMs in typing of the blood group B antigen: Functionality and stability studies. Authors: Tomasz Klaus 1,2, Monika Bzowska 1,2, Małgorzata Kulesza 2,
Agglutinating mouse IgG3 compares favourably with IgMs in typing of the blood group B antigen: Functionality and stability studies. Authors: Tomasz Klaus 1,2, Monika Bzowska 1,2, Małgorzata Kulesza 2,
- PDI5. - Actin A 300-UTR5. PDI5 lines WT. Arabidopsis Chromosome 1. PDI5 (At1g21750) SALK_ SALK_ SALK_015253
 Supplemental Data. Ondzighi et al. (2008). Arabidopsis Protein Disulfide Isomerase-5 Inhibits ysteine Proteases During Trafficking to Vacuoles Prior to Programmed ell Death of the dothelium in Developing
Supplemental Data. Ondzighi et al. (2008). Arabidopsis Protein Disulfide Isomerase-5 Inhibits ysteine Proteases During Trafficking to Vacuoles Prior to Programmed ell Death of the dothelium in Developing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
Supplementary Fig. S1
 Supplementary Fig. S1 CL IP Unc931Flag + + + + IP: αh I: αflg Unc931 CL IP Unc931Flag + + + + FL IP: αflg I: αh Shorter fragment Supplementary Fig. S1: Unc931 associates with full length and Cterminal
Supplementary Fig. S1 CL IP Unc931Flag + + + + IP: αh I: αflg Unc931 CL IP Unc931Flag + + + + FL IP: αflg I: αh Shorter fragment Supplementary Fig. S1: Unc931 associates with full length and Cterminal
Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer.
 Title Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer. Authors H.L. Tan 1, *, C. Yong 1, B.Z. Tan 1, W.J. Fong 1, J. Padmanabhan
Title Conservation of oncofetal antigens on human embryonic stem cells enables discovery of monoclonal antibodies against cancer. Authors H.L. Tan 1, *, C. Yong 1, B.Z. Tan 1, W.J. Fong 1, J. Padmanabhan
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response
 A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
VERIFY Tagged Antigen. Validation Data
 VERIFY Tagged Antigen Validation Data Antibody Validation Figure 1. Over-expression cell lysate for human STAT3 (NM_139276) was used to test 3 commercial antibodies. Antibody A shows strong antigen binding.
VERIFY Tagged Antigen Validation Data Antibody Validation Figure 1. Over-expression cell lysate for human STAT3 (NM_139276) was used to test 3 commercial antibodies. Antibody A shows strong antigen binding.
SUPPLEMENTARY INFORMATION. Tolerance of a knotted near infrared fluorescent protein to random circular permutation
 SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
SUPPLEMENTARY INFORMATION Tolerance of a knotted near infrared fluorescent protein to random circular permutation Naresh Pandey 1,3, Brianna E. Kuypers 2,4, Barbara Nassif 1, Emily E. Thomas 1,3, Razan
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
PCR analysis was performed to show the presence and the integrity of the var1csa and var-
 Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 EPRS expression in multiple cell lines upon viral infection. (a,b) EPRS is slightly induced upon viral induction. qpcr of Eprs mrna (a) and immunoblot analysis of corresponding endogenous
Supplementary Figure 1 EPRS expression in multiple cell lines upon viral infection. (a,b) EPRS is slightly induced upon viral induction. qpcr of Eprs mrna (a) and immunoblot analysis of corresponding endogenous
A guide to selecting control, diluent and blocking reagents
 Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex
 POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
POSTER PRESENTATION Transcriptional regulation of IFN-l genes in Hepatitis C virus-infected hepatocytes via IRF-3 IRF-7 NF- B complex Hai-Chon Lee *, Je-In Youn, Kyungwha Lee, Hwanyul Yong, Seung-Yong
A guide to selecting control, diluent and blocking reagents
 Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
Specializing in Secondary Antibodies and Conjugates A guide to selecting control, diluent and blocking reagents Optimize your experimental protocols with Jackson ImmunoResearch Secondary antibodies and
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
SUPPLEMENTARY MATERIALS Supplementary Table S1: List of primers used for ChIP analysis and Oligo-pull-down assay. Genes Sequence mppargc1a FW 5 -GCGAGGTTTCTGCTTAGTCA-3 (-2317) RV 5 -ACAATGACTAAGCAGAAACCTCG-3
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
B Western blot of the p150caf-1 complex
 Loyola et al., Supplementary Figure 1 A MS data of the HP1α complex RIF1 SPT6 SMARCA4 POGZ CAF-1 p150 KAP-1 HP1α 200 116 97.1 CAF-1 p150 tnasp Importin4 B Western blot of the p150caf-1 complex Input mock
Loyola et al., Supplementary Figure 1 A MS data of the HP1α complex RIF1 SPT6 SMARCA4 POGZ CAF-1 p150 KAP-1 HP1α 200 116 97.1 CAF-1 p150 tnasp Importin4 B Western blot of the p150caf-1 complex Input mock
Description of supplementary material file
 Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
Description of supplementary material file In the supplementary results we show that the VHL-fibronectin interaction is indirect, mediated by fibronectin binding to COL4A2. This provides additional information
Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg
![Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg](/thumbs/88/115416970.jpg) Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence
 PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence Parul Singh 1,2, Rameshwaram Nagender Rao 1, Jala Ram Chandra Reddy 3, R.B.N. Prasad 3, Sandeep
PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence Parul Singh 1,2, Rameshwaram Nagender Rao 1, Jala Ram Chandra Reddy 3, R.B.N. Prasad 3, Sandeep
SUPPLEMENTARY INFORMATION
 ARTICLE NUMBER: 16054 DOI: 10.1038/NMICROBIOL.2016.54 Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2 Kouyuki Hirayasu, Fumiji Saito, Tadahiro Suenaga, Kyoko Shida,
ARTICLE NUMBER: 16054 DOI: 10.1038/NMICROBIOL.2016.54 Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2 Kouyuki Hirayasu, Fumiji Saito, Tadahiro Suenaga, Kyoko Shida,
Executing T-REX in mammalian cells
 Supplementary Figure 1 Executing T-REX in mammalian cells HEK-293 cells cultured (a) in 2x 55 cm 2 adherent cell culture plates, and (b and c) in a 48-well multi-well adherent cell culture plate. No cover
Supplementary Figure 1 Executing T-REX in mammalian cells HEK-293 cells cultured (a) in 2x 55 cm 2 adherent cell culture plates, and (b and c) in a 48-well multi-well adherent cell culture plate. No cover
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary Information
 Supplementary Information Deletion of the B-B and C-C regions of inverted terminal repeats reduces raav productivity but increases transgene expression Qingzhang Zhou 1, Wenhong Tian 2, Chunguo Liu 3,
Supplementary Information Deletion of the B-B and C-C regions of inverted terminal repeats reduces raav productivity but increases transgene expression Qingzhang Zhou 1, Wenhong Tian 2, Chunguo Liu 3,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
mcherry Polyclonal Antibody Catalog Number PA Product data sheet
 Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Polyclonal Antibody Catalog Number PA5-34974 Product data sheet Details Size
Website: thermofisher.com Customer Service (US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 mcherry Polyclonal Antibody Catalog Number PA5-34974 Product data sheet Details Size
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
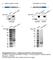 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Human Cell-Free Protein Expression System
 Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
pt7ht vector and over-expressed in E. coli as inclusion bodies. Cells were lysed in 6 M
 Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
SUPPLEMENTARY INFORMATION
 Figure S1 The effect of T198A mutation on p27 stability. a, Hoechst 33342 staining for nuclei (see Fig 1d). Scale bar, 100 μm. b, Densitometric analysis of wild type and mutant p27 protein levels represented
Figure S1 The effect of T198A mutation on p27 stability. a, Hoechst 33342 staining for nuclei (see Fig 1d). Scale bar, 100 μm. b, Densitometric analysis of wild type and mutant p27 protein levels represented
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin
 Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Supplementary Figure 1 (A), (B), and (C) Docking of a physiologic ligand of integrin αvβ3, the tenth type III RGD domain of wild-type fibronectin (A), D1-CD2 (B), and the D1-CD2 variant 3 (C) to Adomain
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Information. A Versatile Tool for Live-Cell Imaging. and Super-Resolution Nanoscopy Studies. of HIV-1 Env Distribution and Mobility
 Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Supplementary Figure 1. Generation of B2M -/- ESCs. Nature Biotechnology: doi: /nbt.3860
 Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Figure 1 Generation of B2M -/- ESCs. (a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III;
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
www.sciencesignaling.org/cgi/content/full/3/146/ra80/dc1 Supplementary Materials for DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination Zhanwen
Results for week 1 Dr Mike Dyall-Smith
 Lecture 2. emm sequence typing of GAS Results for week 1 Dr Mike Dyall-Smith Review of week 1 results Why (sero)group and (sero)type streptococci? Typing schemes: M, SOF, T antigens emm sequence typing
Lecture 2. emm sequence typing of GAS Results for week 1 Dr Mike Dyall-Smith Review of week 1 results Why (sero)group and (sero)type streptococci? Typing schemes: M, SOF, T antigens emm sequence typing
a. Increased active HPSE (50 kda) protein expression upon infection with multiple herpes viruses: bovine
 Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Product Datasheet. LMO2 Antibody NB Unit Size: 0.1 ml
 Product Datasheet LMO2 Antibody NB110-83978 Unit Size: 0.1 ml Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews,
Product Datasheet LMO2 Antibody NB110-83978 Unit Size: 0.1 ml Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews,
SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet
 -Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
-Embryonic Stem Cell Marker (ab15830) 1/8 ページ d Products: Stem Cells >> Germline Stem Cells >> Embryonic Germ Cells SOX2 antibody - Embryonic Stem Cell Marker (ab15830) datasheet Product Name Product type
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative
 Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Table S1. Nucleotide sequences of synthesized oligonucleotides for quantitative RT-PCR For CD8-1 transcript, forward primer CD8-1F 5 -TAGTAACCAGAGGCCGCAAGA-3 reverse primer CD8-1R 5 -TCTACTAAGGTGTCCCATAGCATGAT-3
Supporting Information. Sequence Independent Cloning and Posttranslational. Enzymatic Ligation
 Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Supporting Information Sequence Independent Cloning and Posttranslational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation Wolfgang Ott,,, Thomas Nicolaus,,
Supplementary Online Content
 Supplementary Online Content Scher HI, Lu D, Schreiber NA, et al. Association of on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate
Supplementary Online Content Scher HI, Lu D, Schreiber NA, et al. Association of on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
hours after food deprivation hours after food deprivation
 Figure S.6 protein (fasted / control).2.8.4 p47 p97 Rpt Ufd 3 24 48 hours after food deprivation mrn (fasted / control) C mrn (denervated / control) 2.5 2.5.5 3.5 3 2.5 2.5.5 p97 ** Npl4 Ufd p47 2 3 4
Figure S.6 protein (fasted / control).2.8.4 p47 p97 Rpt Ufd 3 24 48 hours after food deprivation mrn (fasted / control) C mrn (denervated / control) 2.5 2.5.5 3.5 3 2.5 2.5.5 p97 ** Npl4 Ufd p47 2 3 4
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Box 7905, Engineering Building I, North Carolina State University, Raleigh, NC 27695, Phone: Fax:
 Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Product datasheet. ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS)
 Product datasheet info@arigobio.com Package: 1 kit ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS) Component Cat. No. Component Name ARG62820 anti-cd34 antibody [4H11(APG)]
Product datasheet info@arigobio.com Package: 1 kit ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS) Component Cat. No. Component Name ARG62820 anti-cd34 antibody [4H11(APG)]
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Fig. S1 TGF RI inhibitor SB effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of
 Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
