Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
|
|
|
- Nigel Carson
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or the western blot signal for ribosomal protein L9 served as loading controls. The size of the proteins is marked on the right. (-) indicates untreated BL2 cells without any overexpression; LMP2a was tagged with a His-Tag, LMP2b and EBNA_LP (2E and 4E) with a FLAG-Tag, and LMP1 as well as all LMP1-mutants with an HA-Tag and were detected with specific anti-tag antibodies. Positions of molecular weight markers (kda) are indicated on the left. 2
2 Supplementary Figure 2: Uncropped northern blots of vtrna levels in response to expression of EBV-encoded latency III proteins. (a) Northern blot analysis was employed to assess the effects of overexpressing EBV-encoded proteins on the vtrna1-1 levels in different BL2 cell lines. (-) indicate untreated BL2 cells, whereas (evc) indicates the empty vector control. EBV strain B95.8 was used as positive infection control (+ EBV). (b) In the presence of an NF-κB inhibitor northern blot analysis showed no LMP1-dependent up-regulation of vtrna1-1. 3
3 Supplementary Figure 3: LMP1 expression in BL2 cells and its effects on vtrna levels and NF-κB signaling (a) A northern blot analyses showing that LMP1 expression does neither affect vtrna1-2, vtrna1-3, nor vtrna2 levels. As a positive control, vtrna levels in EBV infected BL2 cells (+EBV) are shown. (b) The vtrna1-1 levels in different B cell lines (either EBV infected or not) were assessed by northern blot analysis. Only in cell lines known to express the EBV-encoded LMP1 (ref 1 ) signals for the vtrna1-1 were evident. In (a) and (b) 5.8S rrna served as internal loading control. (c) Real time qpcr of known NF-κB target genes Bcl-xL, IL6 and c-flip was monitored in untreated BL2 cells (-) and in LMP1 expressing cells and compared to cells treated with the NF-κB inhibitor. Fold induction was calculated using the comparative Δct method where BL2 served as calibrator and TBP as housekeeping gene. Data shown are the mean values and standard deviations from three experiments. Significant differences in fold induction relative to the untreated BL2 cells were determined using the two-tailed unpaired Student s t-test (*** P<0.001, ** P<0.01). (d) Western Blot analysis of NF-κB p65 in different BL2 cell lines. GAPDH serves as internal loading control. Positions of molecular weight markers (in kda) are indicated on the left. 4
4 Supplementary Figure 4: (a) Growth curves showing BL41 cells (black) compared to vtrna1-1 overexpressing BL41 cells (red). Cells were counted by a CASY cell counter (Model DT). The mean and standard deviations of three independent proliferation experiments are shown (two-tailed unpaired Student s t-test; * P<0.05). (b) The cell cycle profile of the parental BL41 cell line was compared to BL41 cells ectopically expressing vtrna1-1. Cells were investigated by FACS analysis after a propidium iodide (PI) stain. (c, d) Of note, the vtrna1-1 levels in the overexpressing BL41 cell line could not be further increased by either EBV infection or LMP1 overexpression. However, a sub-clonal cell line of BL41 ectopically expressing vtrna1-1 to a lesser extent (BL41* + vtrna1-1) was responsive to LMP1 expression. This cell line was transfected with the LMP1 expression construct and the levels of LMP1 (c) and vtrna1-1 (d) were monitored by western and northern blot analyses, respectively. In (d) the levels of vtrna1-1 were quantified and compared to the levels seen in the sub-clonal cell line BL41*+vtRNA1-1. In (d) the cell line named BL41 + vtrna1-1 corresponds to the one otherwise used throughout this manuscript. Positions of molecular weight markers (M) are indicated on the left. The ribosomal protein L9 and the 5.8S rrna served as internal loading controls. 5
5 Supplementary Figure 5: Uncropped northern blots assaying the expression levels of human vtrnas. Northern blot analysis reveals the amount of ectopically expressed vtrna1-1 (99 nt long), 1-2 (88 nt long), and 1-3 (89 nt long) in BL41 cells. 6
6 Supplementary Figure 6: Uncropped northern blots to assess the dose dependence of vtrna1-1 levels on the impact of apoptosis resistance. (a) Northern blot analyses demonstrate an shrna-mediated knock-down of vtrna1-1 levels in BL41 cells of ~50%. (b) Northern blot analysis showed a ~60% reduction of endogenous vtrna1-1 levels in HeLa cells upon transfection of an anti-sense oligonucleotide analog (ASO). Administration of a control oligonucleotide (ASO_ctr) strand had no effect. ( ) depict HeLa cells that were treated as the transfected samples, but in the absence of any oligonucleotides. (c) Analogous to (b), vtrna1-1 concentrations were monitored using an anti-sense oligonucleotide analog (ASO) in the human breast cancer cell line HS578T. (d) In the presence of the NF-κB inhibitor IKK VII, northern blot analysis showed a decreased vtrna1-1 expression level in HeLa and HS578T cells. 7
7 Supplementary Figure 7: vtrna1-1 levels affect apoptosis resistance and cell proliferation in BL2 cells. (a) The endogenous vtrna1-1 levels in BL2 cells were further increased by introducing additional vtrna1-1 gene copies during two rounds of lentiviral transfections (+ 2x vtrna1-1). Furthermore an shrnamediated knock-down of vtrna1-1 levels in these cells was performed and the intracellular vtrna1-1 concentrations assessed by northern blot analyses. 5.8S rrna serves as internal loading control. The endogenous levels of vtrna1-1 in untreated BL2 cells (-) was taken as 1.00 and compared to BL2 cells ectopically overexpressing vtrna1-1 (+ 2x vtrna1-1) in the absence or presence of shrnas. (b) BL2 cells without (-) or containing additional vtrna1-1 gene copies were treated with staurosporine (Stau) and the amount of apoptotic cells was determined by flow cytometry analysis after an annexin V stain. The data represent the mean and standard deviations of three independent experiments. The numbers of apoptotic cells in the untreated controls were below 20% and were subtracted from each experiment. (c) Growth curves showing BL2 cells (blue) compared to vtrna1-1 overexpressing BL2 cells (green). The mean and standard deviations of three independent proliferation experiments are shown. P values were determined using the two-tailed unpaired Student s t-test (** P<0.01). 8
8 Supplementary Figure 8: Uncropped northern blots showing expression levels of mutant versions of vtrna. The expression levels of mutant vtrnas M1 to M5 in BL41 were analyzed by northern blot analyses with an oligonucleotide able to detect all vtrna variants. The asterisk depicts a signal from a previous northern blot hybridization using the same nylon membrane but a radiolabeled probe targeting 5.8S rrna. 9
9 Supplementary Figure 9: Uncropped western and northern blots monitoring levels of MVP and vtrna1-1 upon MVP knock-down. (a) MVP levels in BL41 cells treated with the empty vector control (evc) or ectopically expressing vtrna1-1 in the absence (+vtrna1-1) or presence of shrna directed against the MVP were probed by western blot analyses. Positions of molecular weight markers (in kda) are indicated on the left. (b) The expression levels of vtrna1-1 in the cells shown in (a) were assessed by northern blotting. Ribosomal protein L9 served as loading control. 10
10 Supplementary Figure 10: vtrna1-1 modulates both the intrinsic and the extrinsic apoptosis pathway. (a) Uncropped western blots showing the protein levels of Bcl-xL, ARC, and the cleaved caspases Casp-9, and Casp-3 in BL41 cells expressing vtrna1-1 or vtrna1-2 in the absence (-) or presence (+) of staurosporine (Stau). Cleavage of Casp-8 was monitored after Fas-L treatment. Positions of molecular weight markers (in kda) are indicated on the left. (b) Uncropped western blots to monitor the kinetics and levels of phosphorylated IκB expression in BL41 cells, or in BL41 cells expressing vtrna1-1 or vtrna1-2, respectively, as a function of TNFα incubation (in minutes). Hsp90 served as loading control. Molecular weight marker (M) is indicated on the left. 11
11 Supplementary Figure 11: mirna21 is not involved in vtrna1-1 mediated apoptosis resistance. (a) vtrna1-1 expression and/or treatment with staurosporine have no effect on mirna21 levels in BL41 cells. 5.8S rrna serves as loading controls for the shown northern blot. (b) In support of the unchanged mirna21 levels, one of the validated cellular targets for mirna21 (PDCD4, a protein involved in the intrinsic apoptosis pathway) was not affected in the cells under investigation. Ribosomal protein L9 served as internal loading control for the western blot. Positions of molecular weight markers (in kda) are indicated on the left. 12
12 Supplementary Table 1: Quantitative PCR-array of 84 key marker genes involved in apoptosis. Fold changes were calculated using the comparative 2 -(ΔΔct) method where BL41 served as calibrator and BL41 expressing vtrna1-1 as target. The mean of two technical replicates is shown. 13
13 Supplementary Table 2: DNA oligonucleotides used in this study 14
14 Supplementary References 1 Kamranvar, S. A., Gruhne, B., Szeles, A. & Masucci, M. G. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene 26, , doi: /sj.onc (2007). 15
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity]
![3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity] 3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity]](/thumbs/87/97394430.jpg) L S p4 3 D3 76 N S L Ve ct or p4 3 D3 76 N S L Ve ct or A aspase 8 FADD TRAF2 D95-R - + Vector D95L TL I S L D376N T RAIL-R1 T RAIL-R2 D95-R E-y5 [Fluorescence intensity] Supplemental Fig. 1 Different
L S p4 3 D3 76 N S L Ve ct or p4 3 D3 76 N S L Ve ct or A aspase 8 FADD TRAF2 D95-R - + Vector D95L TL I S L D376N T RAIL-R1 T RAIL-R2 D95-R E-y5 [Fluorescence intensity] Supplemental Fig. 1 Different
Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of Bcl-10.
 α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
α-cd3 + α-cd28: Time (min): + + + + + + + + + 0 5 15 30 60 120 180 240 300 360 360 n.s. Supplementary Fig. 1 Kinetics of appearence of the faster migrating form of. Immunoblot of lysates from Jurkat cells
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Fig. S1 TGF RI inhibitor SB effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of
 Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Fig. S1 TGF RI inhibitor SB525334 effectively blocks phosphorylation of Smad2 induced by TGF. FET cells were treated with TGF in the presence of different concentrations of SB525334. Cells were lysed and
Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were. oligonucleotide spanning the NF-kB site (5 -GATCC-
 SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
SUPPLEMENTARY MATERIALS AND METHODS Electrophoretic Mobility Shift Assay (EMSA). Nuclear extracts were prepared as previously described. (1) A [ 32 P] datp-labeled doublestranded oligonucleotide spanning
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
www.sciencesignaling.org/cgi/content/full/10/496/eaam6291/dc1 Supplementary Materials for Regulation of autophagy, NF-κB signaling, and cell viability by mir-124 in KRAS mutant mesenchymal-like NSCLC cells
Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity
 Immunity, Volume 39 Supplemental Information Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity Jorg van Loosdregt, Veerle Fleskens, Juan
Immunity, Volume 39 Supplemental Information Stabilization of the Transcription Factor Foxp3 by the Deubiquitinase USP7 Increases Treg-Cell-Suppressive Capacity Jorg van Loosdregt, Veerle Fleskens, Juan
Supplementary Figure 1 (related to Figure 1) a. SVEC cells stably expressing egfp tbid 2A BCL xl were analysed by Western blot for expression of egfp
 Supplementary Figure 1 (related to Figure 1) a. SVEC cells stably expressing egfp tbid 2A BCL xl were analysed by Western blot for expression of egfp tbid and BCL xl. TOM20 was used as a loading control.
Supplementary Figure 1 (related to Figure 1) a. SVEC cells stably expressing egfp tbid 2A BCL xl were analysed by Western blot for expression of egfp tbid and BCL xl. TOM20 was used as a loading control.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
b alternative classical none
 Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure. 1: Related to Figure.1 a d e b alternative classical none NIK P-IkBa Total IkBa Tubulin P52 (Lighter) P52 (Darker) RelB (Lighter) RelB (Darker) HDAC1 Control-Sh RelB-Sh NF-kB2-Sh
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
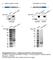 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
supplementary information
 DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
DOI: 10.1038/ncb2116 Figure S1 CDK phosphorylation of EZH2 in cells. (a) Comparison of candidate CDK phosphorylation sites on EZH2 with known CDK substrates by multiple sequence alignments. (b) CDK1 and
 Figure S1: NIK-deficient cells resist to Tweak/TNFα-induced cell death (a) Two independent clones of NIK WT and NIK KO MEFs were treated for 24 hours with Tweak (200 ng/ml) and TNFα (200 U/ml) and cell
Figure S1: NIK-deficient cells resist to Tweak/TNFα-induced cell death (a) Two independent clones of NIK WT and NIK KO MEFs were treated for 24 hours with Tweak (200 ng/ml) and TNFα (200 U/ml) and cell
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
a. Increased active HPSE (50 kda) protein expression upon infection with multiple herpes viruses: bovine
 Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
Fold change vs uninfeced MOI 0.1 MOI 1 MOI 0.1 MOI 1 MOI 0.001 MOI 0.01 Fold change vs uninfected a 6 4 2 50 kda HPSE * ** * * BHV PRV HSV-2 333 ** * 0 - + + - + + - + + b 50 kda HPSE 3 2 ** *** mock inf
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency.
 Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Supplementary Figure 1, related to Figure 1. GAS5 is highly expressed in the cytoplasm of hescs, and positively correlates with pluripotency. (a) Transfection of different concentration of GAS5-overexpressing
Li et al., Supplemental Figures
 Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
Li et al., Supplemental Figures Fig. S1. Suppressing TGM2 expression with TGM2 sirnas inhibits migration and invasion in A549-TR cells. A, A549-TR cells transfected with negative control sirna (NC sirna)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
DOI: 10.1038/ncb3562 In the format provided by the authors and unedited. Supplementary Figure 1 Glucose deficiency induced FH-ATF2 interaction. In b-m, immunoblotting or immunoprecipitation analyses were
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines
 Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines Boris Jakopovich, Ivan Jakopovich, Neven Jakopovich Dr Myko San Health from Mushrooms Miramarska c. 109, Zagreb,
Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines Boris Jakopovich, Ivan Jakopovich, Neven Jakopovich Dr Myko San Health from Mushrooms Miramarska c. 109, Zagreb,
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
Technical Bulletin. Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer. Introduction
 March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma
 Author s response to reviews Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma Authors: Masatoshi Tagawa (mtagawa@chiba-cc.jp)
Author s response to reviews Title: Combination of a third generation bisphosphonate and replication-competent adenoviruses augments the cytotoxicity on mesothelioma Authors: Masatoshi Tagawa (mtagawa@chiba-cc.jp)
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Epstein-Barr Virus Latency in B Cells Leads to Epigenetic Repression and CpG Methylation of the Tumour Suppressor Gene Bim
 Epstein-Barr Virus Latency in B Cells Leads to Epigenetic Repression and CpG Methylation of the Tumour Suppressor Gene Bim Kostas Paschos, Paul Smith, Emma Anderton, Jaap M. Middeldorp, Robert E. White,
Epstein-Barr Virus Latency in B Cells Leads to Epigenetic Repression and CpG Methylation of the Tumour Suppressor Gene Bim Kostas Paschos, Paul Smith, Emma Anderton, Jaap M. Middeldorp, Robert E. White,
Nature Immunology: doi: /ni Supplementary Figure 1. Control experiments for Figure 1.
 Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Supplementary Figure 1 Control experiments for Figure 1. (a) Localization of SETX in untreated or PR8ΔNS1 virus infected A549 cells (4hours). Nuclear (DAPI) and Tubulin staining are shown. SETX antibody
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Cell Proliferation and Death
 Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Transcription Factor Runx3 Is Induced by Influenza A Virus and Double-Strand RNA and
 Transcription Factor Is Induced by Influenza A Virus and Double-Strand RNA and Mediates Airway Epithelial Cell Apoptosis Huachen Gan 1, Qin Hao 1, Steven Idell 1,2 & Hua Tang 1 1 Department of Cellular
Transcription Factor Is Induced by Influenza A Virus and Double-Strand RNA and Mediates Airway Epithelial Cell Apoptosis Huachen Gan 1, Qin Hao 1, Steven Idell 1,2 & Hua Tang 1 1 Department of Cellular
Primers used for PCR of conductin, SGK1 and GAPDH have been described in (Dehner et al,
 Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Description. Lipodin-Pro TM - Protein Transfection Reagent. 1. Kit Benefits
 Description Lipodin-Pro TM - Protein Transfection Reagent The delivery of proteins inside living cells represents an alternative to nucleic acids transfection and a powerful strategy for functional studies
Description Lipodin-Pro TM - Protein Transfection Reagent The delivery of proteins inside living cells represents an alternative to nucleic acids transfection and a powerful strategy for functional studies
REAL TIME PCR USING SYBR GREEN
 REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
FSC-H FSC-H FSC-H
 Supplement Figure A 4 h control + h B 4 h control + h Supplement Figure A-H BV6/TNF-α + ctrl A B 9 h C D h E F 8 h G H Supplement Figure I-P + ctrl I J 8 h K L 4 h M N h O P Supplement Figure 3 A Survival
Supplement Figure A 4 h control + h B 4 h control + h Supplement Figure A-H BV6/TNF-α + ctrl A B 9 h C D h E F 8 h G H Supplement Figure I-P + ctrl I J 8 h K L 4 h M N h O P Supplement Figure 3 A Survival
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
Supplementary Figure 1, Wiel et al
 Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Supplementary Figure 1, Wiel et al Supplementary Figure 1 ITPR2 increases in benign tumors and decreases in aggressive ones (a-b) According to the Oncomine database, expression of ITPR2 increases in renal
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in
 Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Supplementary information Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer s disease Zhentao Zhang, Mingke Song, Xia Liu, Seong Su Kang, Il-Sun Kwon, Duc
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2.
 Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Supplementary Table 1. Primers used to construct full-length or various truncated mutants of ISG12b2. Construct name ISG12b2 (No tag) HA-ISG12b2 (N-HA) ISG12b2-HA (C-HA; FL-HA) 94-283-HA (FL-GFP) 93-GFP
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway
 Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Supplementary Information
 Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
Supplementary Information Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells Kai Fu 1, 6, Xin Sun 1, 6, Wenxin Zheng 1, 6, Eric M. Wier 1, Andrea Hodgson
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Figure S1. Mechanism of ON induced MM cell death.
 Figure S1. Mechanism of ON123300 induced MM cell death. To elucidate whether ON123300 induced decrease of viability of MM cells was associated with induction of apoptosis, the number of apoptotic cells
Figure S1. Mechanism of ON123300 induced MM cell death. To elucidate whether ON123300 induced decrease of viability of MM cells was associated with induction of apoptosis, the number of apoptotic cells
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Nature Structural & Molecular Biology: doi: /nsmb.3308
 Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Figure 1 Analysis of CED-3 autoactivation and CED-4-induced CED-3 activation. (a) Repeat experiments of Fig. 1a. (b) Repeat experiments of Fig. 1b. (c) Quantitative analysis of three independent
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
www.sciencesignaling.org/cgi/content/full/5/233/ra50/dc1 Supplementary Materials for Epidermal Growth Factor Receptor Is Essential for Toll-Like Receptor 3 Signaling Michifumi Yamashita, Saurabh Chattopadhyay,
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Pro-DeliverIN TM - Protein Delivery Reagent Results
 Pro-DeliverIN TM - Protein Delivery Reagent Results OZ Biosciences is delighted to announce the launching of the innovative Pro-DeliverIN - Protein Delivery Reagent. Pro-DeliverIN is a lipid based formulation
Pro-DeliverIN TM - Protein Delivery Reagent Results OZ Biosciences is delighted to announce the launching of the innovative Pro-DeliverIN - Protein Delivery Reagent. Pro-DeliverIN is a lipid based formulation
Supplementary Information. ATM and MET kinases are synthetic lethal with. non-genotoxic activation of p53
 Supplementary Information ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53 Kelly D. Sullivan 1, Nuria Padilla-Just 1, Ryan E. Henry 1, Christopher C. Porter 2, Jihye Kim 3,
Supplementary Information ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53 Kelly D. Sullivan 1, Nuria Padilla-Just 1, Ryan E. Henry 1, Christopher C. Porter 2, Jihye Kim 3,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
B. C. Staurosporine BEZ235 DMSO. -actin. Suppl. Fig. 1 BEZ235 and BGT226 inhibit phosphorylation of Akt and downstream targets of PI3K and mtor.
 DMSO Staurosporine BGT226 A. B. C. BGT226 P-Akt 0 1 2 4 8 hrs. P-Akt 0 5 10 25 50 100 nm PS6 P-4E-BP1 -actin C-Caspase3 -actin PS6 P-4E-BP1 -actin Suppl. Fig. 1 and BGT226 inhibit phosphorylation of Akt
DMSO Staurosporine BGT226 A. B. C. BGT226 P-Akt 0 1 2 4 8 hrs. P-Akt 0 5 10 25 50 100 nm PS6 P-4E-BP1 -actin C-Caspase3 -actin PS6 P-4E-BP1 -actin Suppl. Fig. 1 and BGT226 inhibit phosphorylation of Akt
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Loss of Cul3 in Primary Fibroblasts
 PSU McNair Scholars Online Journal Volume 5 Issue 1 Humans Being: People, Places, Perspectives and Processes Article 15 2011 Loss of Cul3 in Primary Fibroblasts Paula Hanna Portland State University Let
PSU McNair Scholars Online Journal Volume 5 Issue 1 Humans Being: People, Places, Perspectives and Processes Article 15 2011 Loss of Cul3 in Primary Fibroblasts Paula Hanna Portland State University Let
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells.
 Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Supplementary Figure 1. Quantitative RT-PCR experimental validation of CRISPR/Cas9 and sgrnas expression in HEK293A transfected cells. HEK293A cells were transfected with the indicated combinations of
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium
 Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Material for: p53-dependent expression of CXCR5 chemokine receptor in MCF-7 breast cancer cells
 Supplementary Material for: p53-dependent expression of CXCR5 chemokine receptor in MCF-7 breast cancer cells Nikita A Mitkin 1, Christina D Hook 1, Anton M Schwartz 1, Subir Biswas 2, Dmitry V Kochetkov
Supplementary Material for: p53-dependent expression of CXCR5 chemokine receptor in MCF-7 breast cancer cells Nikita A Mitkin 1, Christina D Hook 1, Anton M Schwartz 1, Subir Biswas 2, Dmitry V Kochetkov
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
www.sciencemag.org/cgi/content/full/1154040/dc1 Supporting Online Material for Selective Blockade of MicroRNA Processing by Lin-28 Srinivas R. Viswanathan, George Q. Daley,* Richard I. Gregory* *To whom
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
DOI: 10.1038/ncb2743 Figure S1 stabilizes cellular protein level, post-transcriptionally. (a, b) and DDR1 were RNAi-depleted from HEK.293.-CBG cells. Western blots with indicated antibodies (a). RT-PCRs
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response
 A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective IL-1β-dependent macrophage immune response Ann E. Lin, Federico C. Beasley, Nadia Keller, Andrew Hollands, Rodolfo Urbano, Emily
