Supplementary Material: Peroxisomes protect lymphoma cells from HDAC inhibitor-mediated apoptosis
|
|
|
- Conrad Booker
- 6 years ago
- Views:
Transcription
1 Supplementary Material: Peroxisomes protect lymphoma cells from HDAC inhibitor-mediated apoptosis Michael S Dahabieh 1,2, ZongYi Ha 1,5, Erminia Di Pietro 3,5, Jessica N Nichol 1, Alicia M Bolt 1,4, Christophe Goncalves 1, Daphné Dupéré-Richer 1, Filippa Pettersson 1, Koren K Mann 1,2,4, Nancy E Braverman 3, Sonia V del Rincón *,1, Wilson H Miller Jr. *,1,2 Affiliations: 1 Lady Davis Institute, McGill University 2 Department of Experimental Medicine, McGill University 3 Department of Medical Genetics and Pediatrics, McGill University Health Centre 4 Department of Oncology, McGill University 5 These authors contributed equally to the work. * To whom correspondence should be addressed: wilson.miller@mcgill.ca, sonia.delrincon@mcgill.ca
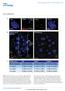
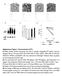
2 Supplementary materials and methods: Mined cdna analyses: Gene set expression profiles corresponding to vehicle and HDACi-treated U937 (GEO: GSE58421), MM231 (GEO: GSE72688), NB4, HL-60, and K562 (GEO: GSE32240) were analyzed with respect to the GO term peroxisome organization. Heat maps of expressed genes only were generated via hierarchical clustering of rows with Euclidian distance as a metric and average linkage method using Morpheus (Broad institute, Cambridge, MA, USA). Phase contrast images: Cells were transfected per the methods section in main text, treated with 2 um Vor for 18h, viewed a DM IL LED (Leica, Wetzlar, Germany) microscope and images were captured via an Infinity3 (Lumenera, Sarasota, FL, USA) camera. Supplementary Figure Captions: Fig. S1. HDACi induce upregulation of peroxisomes across multiple cancer cell lines. (A) Microarray heatmap of cdnas corresponding to GO term: peroxisome organization (GO: ) in Vor-treated (2 µm, 12h) U937 cells (GEO: GSE58421), performed in biological duplicate (N=2). (B) Mined, microarray expression profile of SAHA-treated (5 µm, 24h) MDA- MB-231 cells from N=3 (GEO accession: GSE72688). Gray bars indicate transcripts with undetectable expression. (C) Microarray expression profile (GEO accession: GSE32240) in NB4, K562 and HL-60 leukemia cell lines upon treatment with (1mM, 48h) valproic acid (VPA). Note: values found on GEO correspond to Log 2 of the ratio of VPA vs vehicle treated. (D)
3 Immunoblots of PEX3 and PMP70 (β-actin loading control) in vehicle (DMSO) and Vor-treated (2 µm) HL-60 and (5 µm) MM-231 cells. (E) Total PlsEtn levels (sn-1, PlsEtn), (F) PlsEtn 16:0, (G) 18:0 and (H) 18:1 after vehicle and 6,12, and 18h Vor (2 µm) treatment of U937 cells. Fig. S2. PEX3 knockdown potentiates Vor-induced apoptosis in lymphoma model systems. (A) PMP70 immunofluorescence staining (DAPI nuclear stain) of siscr and sipex3-2 vehicle (DMSO) and Vor-treated (2 µm, 12h) U937 cells. Vor was added 72h post-transfection. Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 with 5 images taken per condition and ~4 cells per image. Scale bar represents 10 µm. * P 0.05 and ***P (B) Immunoblots comparing siscr vs sipex3-2 knockdown in U937 cells following DMSO and Vor (2 µm, 12h) treatment. Shown are additional peroxisome biogenesis and assembly components PEX16 and PEX19, as well as apoptosis marker CL-CASP3. (C) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sipex3-2 upon vehicle and Vor treatment (2 µm, 24h) of U937 cells and (D) corresponding to bar graphs of apoptotic cells representing the sum of Annexin-V (+)/PI(+) and Annexin-V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). *** P (E) Phase contrast cell images corresponding to treatments upon PEX3 knockdown in U937 cells. Scale bar represents 10 µm. (F) Relative PEX3, and CL-CASP3 protein levels upon of sipex3-1 and sipex3-2 transfection and Vor treatment (4 µm, 16h) in OCI-LY8 cells, and (G) Flow cytometry scatter plots of AnnexinV- FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sipex3-1, sipex3-2 upon vehicle and Vor treatment (4 µm, 24h) of OCI-LY8 cells and (H) corresponding bar graphs of apoptotic cells representing the sum of Annexin-V (+)/PI(+) and Annexin-V(+)/PI(-).
4 Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). *** P Fig. S3. PEX19 knockdown potentiates Vor-induced apoptosis in lymphoma model systems. (A) Immunoblot of PEX19 levels upon siscr and sipex19 (2 sequences) transfection following vehicle (DMSO) and Vor (2 µm, 12h) treatment in U937 cells, and (B) OCI-LY8 cells (4 µm, 12h). (C) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sipex3 (sequences 1 and 2) upon vehicle and Vor treatment (2 µm, 24h) of U937 cells and (D) OCI-LY8 cells (4 µm Vor, 24h). (E) and (F) correspond to bar graphs of apoptotic cells from (C) and (D), respectively, representing the sum of Annexin-V (+)/PI(+) and Annexin-V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). ** P 0.01, *** P Fig. S4. B8 cells possess elevated plasmalogen levels and undergo apoptosis upon PEX3 knockdown. Analyses of (A) PlsEtn 18:0, and (B) 16:0 upon vehicle (DMSO) and 12h Vor (2 µm) treatment in U937 cells and B8 cells (2 µm Vor). Graphs represent means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). ** P 0.01, *** P (C) PMP70 immunofluorescence staining (DAPI nuclear stain) of siscr and sipex3-2 vehicle (DMSO) and Vor-maintained B8 cells. Graph represents means ± SEM (unpaired t-test) of N=3 (technical triplicate). ***P Scale bar represents 10 µm. (D) Immunoblots of PEX3, PEX16, PEX19, and β-actin (loading control), 96h post transfection with siscr and sipex3-2 in B8 cells (2 µm Vor). (E) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and
5 propidium iodide (PI, vertical axis), of siscr and sipex3-1 and (F) siscr vs sipex3-2 in Vorcultured B8 cells. Graph represents means ± SEM (unpaired t-test) of N=3 from. ** P = Fig. S5. Catalase protects U937 and B8 cells from H 2 O 2 -mediated apoptosis. (A) quantitation of catalase puncta in vehicle (DMSO) and Vor-treated (2 µm, 12h) U937 cells and B8 cells chronically maintained in 2 µm Vor. Graph represents means ± SEM of N=6 with 5 images taken per condition with ~ 6 cells per image. One-way ANOVA analysis. *** P-value (B) Representative immunoblots of catalase, γh2ax, CL-PARP and b-actin (loading control) upon H 2 O 2 (0.25 mm, 12h) treatment of siscr and sicat-1 transfected B8 cells. (C) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sicat (sequences 1 and 2) upon vehicle and 0.25 mm H 2 O 2 treatment (24h) of U937 cells and (D) B8 cells in 0.50 mm H 2 O 2 (24h). Cells were treated with H 2 O 2 48h posttransfection. (E) and (F) correspond to bar graphs of apoptotic/necrotic cells from (C) and (D), respectively, representing the sum of Annexin-V (-)/PI(+), Annexin-V (+)/PI(+) and Annexin- V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). ** P 0.01, *** P Fig. S6. Knockdown of catalase sensitizes B8 cells to HDACi. (A) Immunoblots of catalase, γh2ax, HO-1, CL-CASP3 and b-actin (loading control), upon siscr and sicat-2 treatment. Samples were harvested 96h post-transfection. (B) Flow cytometry scatter plots of AnnexinV- FITC (horizontal axis) and propidium iodide (PI, vertical axis) of siscr and sicat-2 treated cells, 96h post-transfection, and (C) corresponding % apoptotic cells graphs of PI/AnnexinV-
6 FITC double positive and AnnexinV-FITC positive cells. Graph represents means ± SEM (unpaired t-test) of N=3. * P = Fig. S7. HDACi-mediated apoptosis is potentiated via catalase knockdown in U937 and OCI- LY8 cells. (A) Immunoblots of catalase and β-actin (loading control) 48h post-transfection with siscr and sicat-1 and sicat-2 sequences in U937, and (B) OCI-LY8 cells. (C) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sicat (sequences 1 and 2) upon vehicle and Vor treatment (2 µm, 24h) of U937 cells and (D) OCI-LY8 cells (4 µm Vor, 24h). (E) and (F) correspond to bar graphs of apoptotic cells from (C) and (D), respectively, representing the sum of Annexin-V (+)/PI(+) and Annexin-V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). *** P Fig. S8. Panobinostat increases peroxisomal protein abundance and potentiates apoptosis upon PEX3 knockdown. (A) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis), of siscr and sipex3 (sequences 1 and 2) upon vehicle and pano treatment (40 nm, 24h) of U937 cells and (B) corresponding bar graphs of % apoptotic cells representing the sum of Annexin-V (+)/PI(+) and Annexin-V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). * P 0.01, *** P (C) Immunoblots of catalase, PEX3, PEX19, PEX11B and b-actin (loading control) upon treatment 6 and 12h panobinostat (pano) treatment (40 nm) in U937 cells, and constant maintenance in B8 cells (40 nm, 1 week). (D) Flow cytometry scatter plots of AnnexinV-FITC (horizontal axis) and propidium iodide (PI, vertical axis) corresponding to
7 vehicle (DMSO)-treated U937 and B8 cells, alongside U937 cells treated with 40 nm pano for 48h and B8 cells maintained in 40 nm pano for 1 week. (E) Bar graphs of apoptotic/necrotic cells from (D), representing the sum of Annexin-V (-)/PI(+), Annexin-V (+)/PI(+) and Annexin- V(+)/PI(-). Graph represents means ± SEM (one way analysis of variance, Tukey) of N=3 (technical triplicate). *** P Supplementary Tables: Table S1: KEGG pathways enriched in panobinostat-treated DLBCL responders versus nonresponders. See methods section in manuscript (GSEA) for analyses details. Table S2: MEF2B mutation status does not correlate with enhanced peroxisome gene expression. Analyses were performed from 4 patients with MEF2B mutant status and no correlation was found between elevated expression of KEGG peroxisome transcripts. See methods section in manuscript (GSEA) for analyses details.
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Technical Bulletin. Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer. Introduction
 March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
of Medicine, Zhejiang University, Hangzhou, Zhejiang , China Michigan, 4424B MS-1, 1301 Catherine Street, Ann Arbor, MI 48109, USA.
 Supplemental figure legends: Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells Huiyin Lan 1,2#, Zaiming Tang 1#, Hongchuan Jin 2, and Yi Sun 1,3,4* 1 Institute
Supplemental figure legends: Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells Huiyin Lan 1,2#, Zaiming Tang 1#, Hongchuan Jin 2, and Yi Sun 1,3,4* 1 Institute
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Multiplex Fluorescence Assays for Adherence Cells without Trypsinization
 Multiplex Fluorescence Assays for Adherence Cells without Trypsinization The combination of a bright field and three fluorescent channels allows the Celigo to perform many multiplexed assays. A gating
Multiplex Fluorescence Assays for Adherence Cells without Trypsinization The combination of a bright field and three fluorescent channels allows the Celigo to perform many multiplexed assays. A gating
HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid
 SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTAL MATERIALS AND METHODS Cell culture, transfection and treatments. HeLa cells stably transfected with an empty pcdna3 vector (HeLa Neo) or with a plasmid encoding vmia (HeLa vmia) 1 were cultured
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
Agilent GeneSpring GX 10: Beyond. Pam Tangvoranuntakul Product Manager, GeneSpring October 1, 2008
 Agilent GeneSpring GX 10: Gene Expression and Beyond Pam Tangvoranuntakul Product Manager, GeneSpring October 1, 2008 GeneSpring GX 10 in the News Our Goals for GeneSpring GX 10 Goal 1: Bring back GeneSpring
Agilent GeneSpring GX 10: Gene Expression and Beyond Pam Tangvoranuntakul Product Manager, GeneSpring October 1, 2008 GeneSpring GX 10 in the News Our Goals for GeneSpring GX 10 Goal 1: Bring back GeneSpring
Supplemental Information. Lithocholic Acid Hydroxyamide Destabilizes. Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting. Deubiquitinase USP2a
 Cell Chemical Biology, Volume 24 Supplemental Information Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting Deubiquitinase USP2a Katarzyna Magiera, Marcin Tomala,
Cell Chemical Biology, Volume 24 Supplemental Information Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting Deubiquitinase USP2a Katarzyna Magiera, Marcin Tomala,
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Fig. 5
 Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
Supplementary Fig. 5 Supplemental Figures legends Supplementary Figure 1 (A) Additional dot plots from CyTOF analysis from untreated group. (B) Gating strategy for assessment of CD11c + NK cells frequency
Supporting Information: Core-Shell Nanoparticle-Based Peptide Therapeutics and Combined. Hyperthermia for Enhanced Cancer Cell Apoptosis
 Supporting Information: Core-Shell Nanoparticle-Based Peptide Therapeutics and Combined Hyperthermia for Enhanced Cancer Cell Apoptosis Birju P. Shah a, Nicholas Pasquale a, Gejing De b, Tao Tan b, Jianjie
Supporting Information: Core-Shell Nanoparticle-Based Peptide Therapeutics and Combined Hyperthermia for Enhanced Cancer Cell Apoptosis Birju P. Shah a, Nicholas Pasquale a, Gejing De b, Tao Tan b, Jianjie
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Nature Medicine: doi: /nm.4464
 Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
High accuracy label-free classification of single-cell kinetic states from
 High accuracy label-free classification of single-cell kinetic states from holographic cytometry of human melanoma cells Miroslav Hejna, Aparna Jorapur, Jun S. Song, Robert L. Judson Supplemental Material
High accuracy label-free classification of single-cell kinetic states from holographic cytometry of human melanoma cells Miroslav Hejna, Aparna Jorapur, Jun S. Song, Robert L. Judson Supplemental Material
Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines
 Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines Boris Jakopovich, Ivan Jakopovich, Neven Jakopovich Dr Myko San Health from Mushrooms Miramarska c. 109, Zagreb,
Agarikon.1 and Agarikon Plus Affect Cell Cycle and Induce Apoptosis in Human Tumor Cell Lines Boris Jakopovich, Ivan Jakopovich, Neven Jakopovich Dr Myko San Health from Mushrooms Miramarska c. 109, Zagreb,
Supplemental Information. A Predictive Model for Selective Targeting. of the Warburg Effect through GAPDH. Inhibition with a Natural Product
 Cell Metabolism, Volume 26 Supplemental Information A Predictive Model for Selective Targeting of the Warburg Effect through GAPDH Inhibition with a Natural Product Maria V. Liberti, Ziwei Dai, Suzanne
Cell Metabolism, Volume 26 Supplemental Information A Predictive Model for Selective Targeting of the Warburg Effect through GAPDH Inhibition with a Natural Product Maria V. Liberti, Ziwei Dai, Suzanne
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
INTRODUCTION TO FLOW CYTOMETRY
 DEPARTEMENT BIOZENTRUM INTRODUCTION TO FLOW CYTOMETRY F ACS C ore F acility Janine Zankl FACS Core Facility 3. Dezember 2015, 4pm Cellular Parameters Granulocytes Monocytes Basophils Lymphocytes Neutrophils
DEPARTEMENT BIOZENTRUM INTRODUCTION TO FLOW CYTOMETRY F ACS C ore F acility Janine Zankl FACS Core Facility 3. Dezember 2015, 4pm Cellular Parameters Granulocytes Monocytes Basophils Lymphocytes Neutrophils
Center Drive, University of Michigan Health System, Ann Arbor, MI
 Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
SUPPLEMENTARY INFORMATION
 A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
Reagents and cell culture Mcl-1 gene expression: real-time quantitative RT-PCR In vitro PP2A phosphatase assay Detection of Mcl-1 in vivo
 Reagents and cell culture Antibodies specific for caspase 3, PARP and GAPDH were purchased from Cell Signaling Technology Inc. (Beverly, MA). Caspase inhibitor z-vad-fmk and ROS scavenger N-acetyl-Lcysteine
Reagents and cell culture Antibodies specific for caspase 3, PARP and GAPDH were purchased from Cell Signaling Technology Inc. (Beverly, MA). Caspase inhibitor z-vad-fmk and ROS scavenger N-acetyl-Lcysteine
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing
 Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Actin cap associated focal adhesions and their distinct role in cellular mechanosensing Dong-Hwee Kim 1,2, Shyam B. Khatau 1,2, Yunfeng Feng 1,3, Sam Walcott 1,4, Sean X. Sun 1,2,4, Gregory D. Longmore
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Analysis of Microarray Data
 Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Gene Signature Lab: Exploring integrative LINCS (ilincs) Data and Signatures Analysis Portal & Other LINCS Resources
 Gene Signature Lab: Exploring integrative LINCS (ilincs) Data and Signatures Analysis Portal & Other LINCS Resources Jarek Meller, PhD BD2K-LINCS Data Coordination and Integration Center University of
Gene Signature Lab: Exploring integrative LINCS (ilincs) Data and Signatures Analysis Portal & Other LINCS Resources Jarek Meller, PhD BD2K-LINCS Data Coordination and Integration Center University of
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Nature Genetics: doi: /ng Supplementary Figure 1. DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome.
 Supplementary Figure 1 DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome. (a,b) Quantitative RT-PCR analysis of CD4 + T cells isolated from the peripheral blood of individuals
Supplementary Figure 1 DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome. (a,b) Quantitative RT-PCR analysis of CD4 + T cells isolated from the peripheral blood of individuals
HCT116 SW48 Nutlin: p53
 Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Figure S HCT6 SW8 Nutlin: - + - + p GAPDH Figure S. Nutlin- treatment induces p protein. HCT6 and SW8 cells were left untreated or treated for 8 hr with Nutlin- ( µm) to up-regulate p. Whole cell lysates
Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK)
 SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
Figure S1. Mechanism of ON induced MM cell death.
 Figure S1. Mechanism of ON123300 induced MM cell death. To elucidate whether ON123300 induced decrease of viability of MM cells was associated with induction of apoptosis, the number of apoptotic cells
Figure S1. Mechanism of ON123300 induced MM cell death. To elucidate whether ON123300 induced decrease of viability of MM cells was associated with induction of apoptosis, the number of apoptotic cells
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Knowledge-Guided Analysis with KnowEnG Lab
 Han Sinha Song Weinshilboum Knowledge-Guided Analysis with KnowEnG Lab KnowEnG Center Powerpoint by Charles Blatti Knowledge-Guided Analysis KnowEnG Center 2017 1 Exercise In this exercise we will be doing
Han Sinha Song Weinshilboum Knowledge-Guided Analysis with KnowEnG Lab KnowEnG Center Powerpoint by Charles Blatti Knowledge-Guided Analysis KnowEnG Center 2017 1 Exercise In this exercise we will be doing
Int. J. Mol. Sci. 2016, 17, 1259; doi: /ijms
 S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
S1 of S5 Supplementary Materials: Fibroblast-Derived Extracellular Matrix Induces Chondrogenic Differentiation in Human Adipose-Derived Mesenchymal Stromal/Stem Cells in Vitro Kevin Dzobo, Taegyn Turnley,
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Nature Methods doi: /nmeth Supplementary Figure 1. Screening for cancer stem cell marker(s)
 Supplementary Figure 1 Screening for cancer stem cell marker(s) (a) Flow cytometry analysis of CD133 at increasing times to analysis. Cells were trypsinized, stained, transferred to RPMI medium and then
Supplementary Figure 1 Screening for cancer stem cell marker(s) (a) Flow cytometry analysis of CD133 at increasing times to analysis. Cells were trypsinized, stained, transferred to RPMI medium and then
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Electronic Supplementary Material (ESI) for Journal of Materials Chemistry This journal is The Royal Society of Chemistry 2011.
 Experimental: MTT assay: To determine cell viability the colorimetric MTT metabolic activity assay was used. Hela cells (1 10 4 cells/well) were cultured in a 96-well plate at 37 C, and exposed to varying
Experimental: MTT assay: To determine cell viability the colorimetric MTT metabolic activity assay was used. Hela cells (1 10 4 cells/well) were cultured in a 96-well plate at 37 C, and exposed to varying
Stefano Monti. Workshop Format
 Gad Getz Stefano Monti Michael Reich {gadgetz,smonti,mreich}@broad.mit.edu http://www.broad.mit.edu/~smonti/aws Broad Institute of MIT & Harvard October 18-20, 2006 Cambridge, MA Workshop Format Morning
Gad Getz Stefano Monti Michael Reich {gadgetz,smonti,mreich}@broad.mit.edu http://www.broad.mit.edu/~smonti/aws Broad Institute of MIT & Harvard October 18-20, 2006 Cambridge, MA Workshop Format Morning
Supporting Information
 Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
For detecting mitochondrial membrane potential changes in cells using our proprietary fluorescence probe.
 ab112133 JC-10 Mitochondrial Membrane Potential Assay Kit Flow Cytometry Instructions for Use For detecting mitochondrial membrane potential changes in cells using our proprietary fluorescence probe. This
ab112133 JC-10 Mitochondrial Membrane Potential Assay Kit Flow Cytometry Instructions for Use For detecting mitochondrial membrane potential changes in cells using our proprietary fluorescence probe. This
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Supplementary Material
 Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions Joshua N. Burton 1, Andrew Adey 1, Rupali P. Patwardhan 1, Ruolan Qiu 1, Jacob O. Kitzman 1, Jay Shendure 1 1 Department
NM interference in the flow cytometric Annexin V/Propidium Iodide measurement
 Project: VIGO NM interference in the flow cytometric Annexin V/Propidium Iodide measurement AUTHORED BY: DATE: Cordula Hirsch 20.01.2014 REVIEWED BY: DATE: Harald Krug 10.04.2014 APPROVED BY: DATE: DOCUMENT
Project: VIGO NM interference in the flow cytometric Annexin V/Propidium Iodide measurement AUTHORED BY: DATE: Cordula Hirsch 20.01.2014 REVIEWED BY: DATE: Harald Krug 10.04.2014 APPROVED BY: DATE: DOCUMENT
Cell Proliferation and Death
 Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Cell Proliferation and Death Derek Davies, Cancer Research UK http://www.london-research-institute.org.uk/technologies/120 Proliferation A cell Apoptosis Cell death Proliferation signals Senescence DNA
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supporting information. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo
 Supporting information Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo Greg Thurber 1, Katy Yang 1, Thomas Reiner 1, Rainer Kohler 1, Peter Sorger 2, Tim Mitchison
Supporting information Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo Greg Thurber 1, Katy Yang 1, Thomas Reiner 1, Rainer Kohler 1, Peter Sorger 2, Tim Mitchison
mir-24-mediated down-regulation of H2AX suppresses DNA repair
 Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplemental Online Material mir-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells Ashish Lal 1,4, Yunfeng Pan 2,4, Francisco Navarro 1,4, Derek M. Dykxhoorn
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Xfect Protein Transfection Reagent
 Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Xfect Protein Transfection Reagent Mammalian Expression Systems Rapid, high-efficiency, low-toxicity protein transfection Transfect a large amount of active protein Virtually no cytotoxicity, unlike lipofection
Orflo Application Note 8 /2017 Rapid Apoptosis Monitoring Using Annexin V and Orflo s Moxi GO II Next Generation Flow Cytometer
 Introduction/Background Orflo Application Note 8 /2017 Cellular apoptosis is a sophisticated mechanism employed by cells to carefully control death in response to cell injury. Commonly referred to as programmed
Introduction/Background Orflo Application Note 8 /2017 Cellular apoptosis is a sophisticated mechanism employed by cells to carefully control death in response to cell injury. Commonly referred to as programmed
Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41590-017-0001-2 In the format provided by the authors and unedited. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41590-017-0001-2 In the format provided by the authors and unedited. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Supplemental Information. A Versatile Tool for Live-Cell Imaging. and Super-Resolution Nanoscopy Studies. of HIV-1 Env Distribution and Mobility
 Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Cell Chemical Biology, Volume 24 Supplemental Information A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility Volkan Sakin, Janina Hanne,
Supplemental Information Inventory
 Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin
 Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
The microtubule-associated tau protein has intrinsic acetyltransferase activity. Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and
 SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
SUPPLEMENTARY INFORMATION: The microtubule-associated tau protein has intrinsic acetyltransferase activity Todd J. Cohen, Dave Friedmann, Andrew W. Hwang, Ronen Marmorstein and Virginia M.Y. Lee Cohen
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Figure 1 BALB/c LYVE1-deficient mice exhibited reduced lymphatic trafficking of all DC subsets after oxazolone-induced sensitization. (a) Schematic overview of the mouse skin oxazolone contact
Supplementary Text. eqtl mapping in the Bay x Sha recombinant population.
 Supplementary Text eqtl mapping in the Bay x Sha recombinant population. Expression levels for 24,576 traits (Gene-specific Sequence Tags: GSTs, CATMA array version 2) was measured in RNA extracted from
Supplementary Text eqtl mapping in the Bay x Sha recombinant population. Expression levels for 24,576 traits (Gene-specific Sequence Tags: GSTs, CATMA array version 2) was measured in RNA extracted from
Whole Transcriptome Analysis of Illumina RNA- Seq Data. Ryan Peters Field Application Specialist
 Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
a Beckman Coulter Life Sciences: White Paper
 a Beckman Coulter Life Sciences: White Paper CytoFLEX Instrument Evaluation Using Biological Specimens Authors: James Tung 1, Dan Condello 3, Albert Donnenberg 4, Erika Duggan 3, Jesus Lemus 1, John Nolan
a Beckman Coulter Life Sciences: White Paper CytoFLEX Instrument Evaluation Using Biological Specimens Authors: James Tung 1, Dan Condello 3, Albert Donnenberg 4, Erika Duggan 3, Jesus Lemus 1, John Nolan
supplementary information
 DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Nature Methods: doi: /nmeth Supplementary Figure 1. Validation of RaPID with EDEN15
 Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
Microarray Technique. Some background. M. Nath
 Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Microarray Technique Some background M. Nath Outline Introduction Spotting Array Technique GeneChip Technique Data analysis Applications Conclusion Now Blind Guess? Functional Pathway Microarray Technique
Supplemental Materials and Methods
 Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Introduction to Bioinformatics. Fabian Hoti 6.10.
 Introduction to Bioinformatics Fabian Hoti 6.10. Analysis of Microarray Data Introduction Different types of microarrays Experiment Design Data Normalization Feature selection/extraction Clustering Introduction
Introduction to Bioinformatics Fabian Hoti 6.10. Analysis of Microarray Data Introduction Different types of microarrays Experiment Design Data Normalization Feature selection/extraction Clustering Introduction
Supplementary Figure legends. Supplementary Methods
 Supplementary Methods Transmission electron microscopy. For transmission electron microscopy (TEM), the cell populations were rinsed with 0.1 Sorensen s buffer (ph 7.5), fixed in 2.5% glutaraldehyde for
Supplementary Methods Transmission electron microscopy. For transmission electron microscopy (TEM), the cell populations were rinsed with 0.1 Sorensen s buffer (ph 7.5), fixed in 2.5% glutaraldehyde for
Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Investigating Apoptosis. For more information:
 Investigating Apoptosis For more information: www.invitrogen.com/flowcytometry/ Flow Cytometry: It s in our DNA October 2, 2009 2009 Invitrogen Corporation. All Rights Reserved. Proprietary and Confidential.
Investigating Apoptosis For more information: www.invitrogen.com/flowcytometry/ Flow Cytometry: It s in our DNA October 2, 2009 2009 Invitrogen Corporation. All Rights Reserved. Proprietary and Confidential.
Supplemental Material for
 Supplemental Material for TXI TELNGIECTSI MUTTED (TM)-MEDITED DN DMGE RESPONSE IN OXIDTIVE STRESS-INDUCED VSCULR ENDOTHELIL CELL SENESCENCE Hong Zhan 1, Toru Suzuki 1,2, Kenichi izawa 1, Kiyoshi Miyagawa,
Supplemental Material for TXI TELNGIECTSI MUTTED (TM)-MEDITED DN DMGE RESPONSE IN OXIDTIVE STRESS-INDUCED VSCULR ENDOTHELIL CELL SENESCENCE Hong Zhan 1, Toru Suzuki 1,2, Kenichi izawa 1, Kiyoshi Miyagawa,
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
www.sciencesignaling.org/cgi/content/full/4/157/ra4/dc1 Supplementary Materials for Genome-Wide RNAi Screen Reveals Disease-Associated Genes That Are Common to Hedgehog and Wnt Signaling Leni S. Jacob,
For knockdown of individual genes, the following TRC library clones were used: Gene Species Library TRC number Designation in manuscript
 Supplemental Methods shrna constructs for validation experiments For knockdown of MLL-AF9, a custom shrna with the following stem sequence directed against the fusion breakpoint (underlined) was designed
Supplemental Methods shrna constructs for validation experiments For knockdown of MLL-AF9, a custom shrna with the following stem sequence directed against the fusion breakpoint (underlined) was designed
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Engineering Nanomedical Systems. Assessing the efficacy of nanomedical therapies
 BME 626 October 9, 2014 Engineering Nanomedical Systems Lecture 12 Assessing the efficacy of nanomedical therapies James F. Leary, Ph.D. SVM Endowed Professor of Nanomedicine Professor of Basic Medical
BME 626 October 9, 2014 Engineering Nanomedical Systems Lecture 12 Assessing the efficacy of nanomedical therapies James F. Leary, Ph.D. SVM Endowed Professor of Nanomedicine Professor of Basic Medical
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1
 number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
Gene expression: Microarray data analysis. Copyright notice. Outline: microarray data analysis. Schedule
 Gene expression: Microarray data analysis Copyright notice Many of the images in this powerpoint presentation are from Bioinformatics and Functional Genomics by Jonathan Pevsner (ISBN -47-4-8). Copyright
Gene expression: Microarray data analysis Copyright notice Many of the images in this powerpoint presentation are from Bioinformatics and Functional Genomics by Jonathan Pevsner (ISBN -47-4-8). Copyright
Comparative Genomic Hybridization
 Comparative Genomic Hybridization Srikesh G. Arunajadai Division of Biostatistics University of California Berkeley PH 296 Presentation Fall 2002 December 9 th 2002 OUTLINE CGH Introduction Methodology,
Comparative Genomic Hybridization Srikesh G. Arunajadai Division of Biostatistics University of California Berkeley PH 296 Presentation Fall 2002 December 9 th 2002 OUTLINE CGH Introduction Methodology,
Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison. CodeLink compatible
 Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison CodeLink compatible Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood
Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood - a Platform Comparison CodeLink compatible Microarray Analysis of Gene Expression in Huntington's Disease Peripheral Blood
Cellometer Vision CBA
 Features of the Vision CBA Image Cytometry System All-in-One System Basic cell counting, primary cell viability, and cellbased assays. See for Yourself Why the Top Ten Pharmaceutical Companies Trust Cellometer
Features of the Vision CBA Image Cytometry System All-in-One System Basic cell counting, primary cell viability, and cellbased assays. See for Yourself Why the Top Ten Pharmaceutical Companies Trust Cellometer
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
sirna delivery systems: lipoplexes vs. polyplexes
 sirna delivery systems: lipoplexes vs. polyplexes Preeti Yadava College of Pharmacy, University of Florida, Gainesville, Florida GPEN 2006 What is RNA interference? RNAi is the natural process of sequencespecific,
sirna delivery systems: lipoplexes vs. polyplexes Preeti Yadava College of Pharmacy, University of Florida, Gainesville, Florida GPEN 2006 What is RNA interference? RNAi is the natural process of sequencespecific,
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
