Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
|
|
|
- Joseph Goodman
- 6 years ago
- Views:
Transcription
1 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr F 5' TTTGCCAAGGCACGAGTAACA 3' 14 bp R 5' ATTCCCAAGGACCACCTCACA 3' hil 6 F 5' AGCCACTCACCTCTTCAGAACGAA 3' 121 bp R 5' AGTGCCTCTTTGCTGCTTTCACAC 3' mil 6 F 5' ATCCAGTTGCCTTCTTGGGAC 3' 134 bp R 5' TAAGCCTCCGACTTGTGAAGT 3' hlif F 5' TATCACCATCTGTGCCTTTGCTGC 3' 187 bp R 5' TCTGCCAGATTGTTCCTATGCCCA 3' mlif F 5' ATACAGCCCAGCACCAGCTAGAAA 3' 138 bp R 5' GCTTGTTCATACACCCAGCAAGCA 3' hgapdh F 5' CCACATGGCCTCCAAGGAGTAAGAC 3' 131 bp R 5' AGGAGGGGAGATTCAGTGTGGTGGG 3' mgapdh F R h:human; m:mouse 5' CTGACGTGCCGCCTGGAGAAA 3' 5' AGCCCCGGCATCGAAGGTGG 3' 165 bp
2 Supplementary Figure 1: Orthotopic co-injection of U87wt and U87Δ cells results in reduced survival. (a-c) Survival analysis after intracranial injection of U87wt (wt), U87Δ (Δ) or U87wt mixed with U87Δ at indicated ratios (wt + Δ) (1%= 5x1 5 cells). (d) Survival analysis summary table indicating Kruskal-Wallis p values. (e) H&E staining of brain sections obtained at day 12 after intracranial injection of U87DK (DK) 1%, U87Δ (Δ) 1% or U87DK mixed with U87Δ at a ratio of 9:1 respectively (1%= 5x1 5 cells). (f) X-Gal stain quantification of intracranial tumors obtained at day 22 after injection of U87wt cells or U87Δ alone or U87wt + U87Δ-LacZ, at a 9:1 ratio, respectively. Supplementary Figure 2: U87DK cells do not enhance U87wt tumorigenicity after intracranial injection. H&E staining of brain sections obtained 14 days after intracranial injection into nude mice of U87wt cells mixed with U87DK at a ratio of 9:1. Supplementary Figure 3: Tumor growth kinetics of mixed glioma cell injections. (a) Tumor growth kinetics after subcutaneous injection into nude mice of U87Δ or U87wt cells injected alone or co-injected with U87Δ, U87DK or U87Par cells. (b) Detailed view of tumor growth kinetics from the lower part of the graph. Error bars represent mean ± s.e.m. Supplementary Figure 4: Orthotopic co-injection of mouse Ink4/Arf -/- -astrocytes engineered with wtegfr and ΔEGFR. (a) H&E of mouse brains 16 or 19 days after intracranial injection of mastr-ink4/arf -/- astrocytes engineered with wtegfr (mastr-wt), ΔEGFR (mastr-δ), or co-injection of mastr-wt plus mastr-δ mixed at a 9:1 ratio, respectively (mastr-wt+δ) (1%= 5x1 5 cells). (b) H&E staining of brain sections after injection of mastr-δ 1% (Δ 1%) or mastr-wt cells mixed with mastr-δ at a ratio of 9:1 respectively (wt + Δ 9-1%). High magnification illustrating perivascular invasive nature of tumor cells (1%= 5x1 5 cells). Supplementary Figure 5: wtegfr and ΔEGFR cells tend to establish equilibrium in vivo. (a) Tumor volume at day 22 after subcutaneous injection of U87wt-LacZ or U87Δ 1% alone, or mixed at ratios 9:1, 5:5 or 1:9 respectively (1%= 2x1 5 cells). (b) Proportion representation of U87wt cells at the injection time and at the end of the experiment (day 22). (c) Analysis of tumor composition by flow cytometry using Ab-1 (FITC) and Ab-5 (APC) antibodies to stain tumors formed after subcutaneous injection of U87wt mixed with U87Δ at a ratio of 9:1. Left, example of tumor composition analysis by flow cytometry. Right, composition of the mixed tumors according to the results obtained by flow cytometry. Error bars represent mean ± s.e.m.
3 Supplementary Figure 6: Analysis of EGFR and STAT3 phosphorylation by western blot analysis of tumor lysates obtained after subcutaneous injection of indicated mixtures of U87 cell line derivatives at indicated ratios. Each lane represents a different tumor sample. Supplementary Figure 7: ΔCM treatment activates wtegfr and downstream pathways in U178 and U373 glioma cells. (a) Analysis of EGFR (pegfr) and STAT3 (pstat3) phosphorylation by western blot of U178wt lysates non treated (-) or treated with U178Δ CM (ΔCM), or U178Δ CM pre-treated with anti-il-6 (ΔCM+IL-6n) or anti-gp13 (ΔCM+gp13n) neutralizing antibodies. (b) Analysis of EGFR (pegfr) and STAT3 (pstat3) phosphorylation by western blot of U373wt lysates non treated (-) or treated with U373Δ CM (ΔCM), or U373Δ CM pre-treated with anti-lif (ΔCM+LIFn) or anti-gp13 (ΔCM+gp13n) neutralizing antibodies. Total protein and actin were used as loading control in all experiments. Supplementary Figure 8: Quantification of EGF, TGF-α, amphiregulin (AR), HB-EGF and betacellulin (BTC) by ELISA in supernatants of U87Par, U87wt, U87Δ and U87DK cells. Error bars represent mean ± s.e.m. Supplementary Figure 9: Quantification of IL-6 and LIF in U178 and U373 glioma cells. Real-time PCR for IL-6 (a) and LIF (b) expression in U178 and U373 (Par) or engineered to over-express wtegfr (wt), ΔEGFR (Δ) or ΔEGFR with a dead kinase domain (DK). (c) ELISA quantification of IL-6 in supernatants of U178 and U373 cell line derivatives after 48 h of starvation. Error bars in all experiments represent mean ± s.e.m. One-way ANOVA and two-tail t-test were used for statistical analysis. **p<.1. Supplementary Figure 1: Quantification of IL-6 family cytokines in U87 cell line derivatives. Real time PCR of the IL-6 family members Cardiotrophin-like cytokine-1 (CLC- 1), Ciliary Neurotrophic Factor (CTNF), Cardiotrophin-1 (CT-1), IL-6. IL-11, IL-27, Leukemia Inhibitory Factor (LIF), IL-11, and Oncostatin M (OSM), in U87Par (Par), U87wt (wt), U87Δ (Δ) and U87DK (DK). Error bars represent mean ± s.e.m. One-way ANOVA and two-tail t-test were used for statistical analysis. **p<.1. Supplementary Figure 11: Blockade of STAT3 activation by IL-6 and LIF neutralizing antibodies. (a) Western blot analysis of EGFR (pegfr) and STAT3 (pstat3) phosphorylation in U87wt lysates non-treated (-), treated with recombinant IL-6 or with U87Δ CM (ΔCM) pre-treated or not with anti-il-6 neutralizing antibody (IL-6n). (b) Western blot analysis of EGFR (pegfr) and STAT3 (pstat3) phosphorylation in U87wt lysates nontreated (-), treated with recombinant LIF or with U87Δ CM (ΔCM) pre-treated or not with anti- LIF neutralizing antibody (LIFn).
4 Supplementary Figure 12: sirna knock-down of gp13, IL-6, and LIF (c) in vitro. (a) Quantification of gp13 levels by ELISA in lysates of U87wt cells non transfected (Neg) or transfected with 25 nm of a negative control sirna against luciferase (Luc sirna) or two different sirnas against gp13 (gp13 sirna#1 and gp13 sirna#2). (b) ELISA quantification of IL-6 secretion in U87Δ cells supernatants transfected with indicated concentrations of a negative control sirna against luciferase (Neg) or two different sirnas against IL-6 (IL6 sirna#1 and IL6 sirna#2). (c) Relative LIF expression after transfection with 25 nm of luciferase sirna (Luc) or LIF sirna by real-time PCR. cdna from non transfected cells was used as a second negative control. Normalization with GAPDH expression was performed. Error bars in all experiments represent mean ± s.e.m. Supplementary Figure 13: sirna knock-down of gp13, IL-6, and LIF in vivo. (a) Tumor growth kinetics after subcutaneous injection into nude mice of U87wt transfected with luciferase sirna (wt-luc sirna) or sirna targeting gp13 (wt-gp13 sirna) alone or mixed with U87Δ at a ratio 9:1 respectively. (b) Tumor growth kinetics after subcutaneous injection into nude mice of U87wt cells injected alone or with U87Δ transfected with luciferase sirna (Δ-Luc sirna) or sirna targeting IL-6 (Δ-IL6-siRNA). (c) Tumor growth kinetics after subcutaneous injection into nude mice of U87wt cells injected alone or with U87Δ transfected with luciferase sirna (Δ-Luc sirna) or sirna targeting LIF (Δ-LIF sirna). Error bars represent mean ± s.e.m. Supplementary Figure 14: Analysis of wtegfr and IL-6 (top) or LIF (bottom) expression by real-time PCR in 19 human GBM samples. Error bars in all experiments represent mean ± s.e.m.
5 Inda_Suppl.Fig1 a b 12% 1% wt 1% 1% 12% 1% wt 1% 1% wt % % survival 8% 6% 4% wt + 9-1% % survival 8% 6% 4% 2% 2% % days after IC injection % days after IC injection c d % survival 12% 1% 8% 6% 4% 2% wt 1%,1% wt + 99,9-,1% Group U87wt 1% 38 Median survival (days) p value 1 p value 2 U87delta 1% 24,7547 U87wt+U87delta 9-1% 23 <,1 U87delta 1% 27 *,482 U87wt+U87delta 99-1% 24,5 <,1 U87delta,1% 61,5 *,114 U87wt+U87delta 99,9-,1% 3 <,1 p value 1 : comparison between U87delta and U87wt+U87wt+delta; p value2: comparison between U87wt and U87wt+U87wt+delta % e days after IC injection DK1% -LacZ 1% DK+ -LacZ 9-1% f 12% 1% % of tumor area 8% 6% 4% 2% LacZ negative LacZ positive % wt 1% 1% wt + 9-1%
6 Inda_Suppl. Fig 2 U87wt 1% U87DK1% U87wt + U87DK 9-1%
7 a 22 Inda_Suppl. Fig 3 Tumor volume (mm 3 ) WT 1% -LacZ 1% -LacZ 1% WT + -LacZ 9-1% -LacZ 1% WT + -LacZ 99-1% WT + DK-LacZ 9-1% WT + DK-LacZ 99-1% WT + Parental-LacZ 9-1% Days post-sq injection b 1 WT 1% Tumor volume (mm3) WT + DK-LacZ 9-1% WT + DK-LacZ 99-1% WT + Parental-LacZ 9-1% Days post-sq injection
8 a Inda_Suppl.Fig 4 mastr-wt 1% mastr-wt+ 9-1% mastr- 1% Day 16 Day 19 b 1% wt + 9-1% 4X 4X 2X
9 a b Inda_Suppl. Fig % Tumor volume (mm 3 ) % of tumor 1% 8% 6% 4% wt 1% wt + 9-1% wt + 5-5% wt + 1-9% 2% % injection end c Δ wt Tumor volume (mm 3 ) wt U87wt 1% U87 1% U87wt + U87 9-1%
10 Inda_Suppl. Fig 6 wt pegfr EGFR wt pstat3 STAT3 Actin
11 Inda_Suppl. Fig 7 a b pegfr pegfr EGFR pakt Akt perk ERK pstat3 STAT3 Actin EGFR pakt Akt perk ERK pstat3 STAT3 Actin
12 Inda_Suppl. Fig 8
13 Inda_Suppl. Fig 9 a Relative expression U178 ** b Relative expression U178 Par wt DK Par wt DK Relative expression 4 2 U373 Par wt DK Relative expression U373 ** Par wt DK c pg/ml U178 ** Par wt DK pg/μl U373 Par wt DK
14 Inda_Suppl. Fig 1 Relative expression Par wt DK ** ** 5 CLC-1 CNTF-1 CT-1 IL-6 IL-11 IL-27 LIF OSM
15 Inda_Suppl. Fig 11 a b Stim.: Ab: - - IL-6 - IL-6 CM CM Stim.: IL-6n - IL-6n Ab: - - LIF - CM - LIF LIFn CM LIFn pegfr pegfr EGFR EGFR pstat3 pstat3 STAT3 STAT3 Actin
16 a 1 Inda_Suppl. Fig 12 pg/5 μg lysate b Neg Luc sirna gp13 sirna#1 gp13 sirna#2 [IL-6] (pg/ml) nM 2nM 4nM.8nM Luc sirna IL-6 sirna#1 IL-6 sirna#2 c 1,2 Relative expression 1,8,6,4,2 (-) Luc sirna LIF sirna
17 Inda_Suppl. Fig 13 a 5 tumor volume (mm3) wt-luc sirna 9% wt-gp13 sirna 9% 1% wt-luc sirna + 9-1% wt-gp13 sirna + 9-1% b c tumor volume (mm 3 ) Tumor volume (mm3) days post SQ wt 9% -Luc sirna 1% -IL6siRNA wt + -Luc sirna 9-1% wt + -IL6 sirna 9-1% days post SQ wt 9% -Luc sirna 1% -LIF sirna 1% wt + -Luc sirna 9-1% wt + -LIF sirna 9-1% days post SQ
18 Inda_Suppl. Fig 14 a nº of human GBM wtegfr + wtegfr - IL-6 high IL-6 low nº of human GBM wtegfr + wtegfr - LIF high LIF low
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets.
 Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Supplementary Figure 1. Characterization of EVs (a) Phase-contrast electron microscopy was used to visualize resuspended EV pellets. Scale bar represent 100 nm. The sizes of EVs from MDA-MB-231-D3H1 (D3H1),
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by. Regulating Occludin
 Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Intestinal Epithelial Cell-Specific Deletion of PLD2 Alleviates DSS-Induced Colitis by Regulating Occludin Chaithanya Chelakkot 1,ǂ, Jaewang Ghim 2,3,ǂ, Nirmal Rajasekaran 4, Jong-Sun Choi 5, Jung-Hwan
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse
 Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Supplementary Figure 1. Bone density was decreased in osteoclast-lineage cell specific Gna13 deficient mice. (a-c) PCR genotyping of mice by mouse tail DNA. Primers were designed to detect Gna13-WT/f (~400bp/470bp)
Initial genotyping of all new litters was performed by Transnetyx (Memphis,
 SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Figure Legend
 Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplementary Figure Legend Supplementary Figure S1. Effects of MMP-1 silencing on HEp3-hi/diss cell proliferation in 2D and 3D culture conditions. (A) Downregulation of MMP-1 expression in HEp3-hi/diss
Supplemental Information. Mitophagy Controls the Activities. of Tumor Suppressor p53. to Regulate Hepatic Cancer Stem Cells
 Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK)
 SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
SUPPLEMENTAL MATERIAL Supplemental Methods Flow cytometry Cell were phenotyped using FITC-conjugated anti-human CD3 (Pharmingen, UK) and anti-human CD68 (Dako, Denmark), anti-human smooth muscle cell α-actin
B Vehicle 1V270 (35 μg) 1V270 (100 μg) Days post tumor implantation. Vehicle 100μg 1V270 biweekly 100μg 1V270 daily
 Supplemental Figure 1 A 1 1V7 (8 μg) 1 1V7 (16 μg) 8 6 4 B 1 1 8 6 4 1V7 (35 μg) 1V7 (1 μg) 5 1 15 5 1 15 C Biweekly 8 11 14 17 Days Implant SCC7 cells Daily 8 9 1 11 1 Implant SCC7 cells 1V7 i.t. treatment
Supplemental Figure 1 A 1 1V7 (8 μg) 1 1V7 (16 μg) 8 6 4 B 1 1 8 6 4 1V7 (35 μg) 1V7 (1 μg) 5 1 15 5 1 15 C Biweekly 8 11 14 17 Days Implant SCC7 cells Daily 8 9 1 11 1 Implant SCC7 cells 1V7 i.t. treatment
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
* ** ** * IB: p-p90rsk. p90rsk (Ser380) (arbitrary units) (Ser380) p90rsk. IB: p90rsk. Tubulin. IB: Tubulin. Ang II (200 nm) Ang II (200 nm)
 I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
I: p-p9rsk I: p9rsk I: C I: p-p9rsk I: p9rsk 5 (ka) 5 5 (min) Ang II ( nm) p-p9rsk (Ser8) p9rsk p-p9rsk (Ser8) p9rsk (h) Mannitol 5 mm -Glucose 5 mm p9rsk (Ser8) (arbitrary units) p-p9rsk (Ser8) (arbitrary
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Meso Scale Discovery Applications
 A Suite of Assays to Detect hosphorylated Receptor Tyrosine Kinases Associated with Neoplasia Timothy S. Schaefer, Jane Zhang, Sean Higgins, Laura K. Schaefer, Robert M. Umek and Jacob N. Wohlstadter Reversible
A Suite of Assays to Detect hosphorylated Receptor Tyrosine Kinases Associated with Neoplasia Timothy S. Schaefer, Jane Zhang, Sean Higgins, Laura K. Schaefer, Robert M. Umek and Jacob N. Wohlstadter Reversible
SBH Sciences. Innovative Biotechnology Company with Two Discovery Programs. Discovery and Pre-clinical Contract Research Organization (CRO)
 SBH Sciences Innovative Biotechnology Company with Two Discovery Programs Discovery and Pre-clinical Contract Research Organization (CRO) Production and Bioassay Testing of Cytokines & Enzymes Bio-Incubator
SBH Sciences Innovative Biotechnology Company with Two Discovery Programs Discovery and Pre-clinical Contract Research Organization (CRO) Production and Bioassay Testing of Cytokines & Enzymes Bio-Incubator
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Real-time PCR. TaqMan Protein Assays. Unlock the power of real-time PCR for protein analysis
 Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
Real-time PCR TaqMan Protein Assays Unlock the power of real-time PCR for protein analysis I can use my real-time PCR instrument to quantitate protein? Do protein levels correlate with related mrna levels
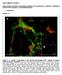
Wnt16 smact merge VK/AB
 A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
A WT Wnt6 smact merge VK/A KO ctrl IgG WT KO Wnt6 smact DAPI SUPPLEMENTAL FIGURE I: Wnt6 expression in MGP-deficient aortae. Immunostaining for Wnt6 and smooth muscle actin (smact) in aortae from 7 day
Molecular analysis and biological implications of STAT3 signal transduction Schuringa, Jan Jacob
 University of Groningen Molecular analysis and biological implications of STAT3 signal transduction Schuringa, Jan Jacob IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
University of Groningen Molecular analysis and biological implications of STAT3 signal transduction Schuringa, Jan Jacob IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
Supplementary Figure 1 Activated B cells are subdivided into three groups
 Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Supplementary Figure 1 Activated B cells are subdivided into three groups according to mitochondrial status (a) Flow cytometric analysis of mitochondrial status monitored by MitoTracker staining or differentiation
Table S1. Primer sequences
 Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Table S1. Primer sequences Primers for quantitative PCR Tgf 1 Forward Tgf 1 Reverse Tgf 2Forward Tgf 2Reverse Tgf 3 Forward Tgf 3 Reverse Tgf r1 Forward Tgf r1 Reverse Tgf r2 Forward Tgf r2 Reverse Thbs1
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
1,500 1,000. LPS + alum. * * Casp1 p10. Casp1 p45
 a NLRP3 Non-stimulation R46 BAY SI TAT LPS R46 BAY SI TAT 1,5 1, c 15 1 5 5 Pro-IL-18 Pro-IL-1 LPS + alum d e f IL-1 p17 Pro-IL-1 1 75 5 5 Casp1 p1 NLRP3 LPS + alum Supplementary Figure 1 Inhiition of
a NLRP3 Non-stimulation R46 BAY SI TAT LPS R46 BAY SI TAT 1,5 1, c 15 1 5 5 Pro-IL-18 Pro-IL-1 LPS + alum d e f IL-1 p17 Pro-IL-1 1 75 5 5 Casp1 p1 NLRP3 LPS + alum Supplementary Figure 1 Inhiition of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
Supplemental Information Inventory
 Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Center Drive, University of Michigan Health System, Ann Arbor, MI
 Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Leukotriene B 4 -induced reduction of SOCS1 is required for murine macrophage MyD88 expression and NFκB activation Carlos H. Serezani 1,3, Casey Lewis 1, Sonia Jancar 2 and Marc Peters-Golden 1,3 1 Division
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation
 1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
1 2 3 4 5 SUPPLEMENTAL DATA Thyroid peroxidase gene expression is induced by lipopolysaccharide involving Nuclear Factor (NF)-κB p65 subunit phosphorylation Magalí Nazar, Juan Pablo Nicola, María Laura
Supplementary Material. Levels of S100B protein drive the reparative process in acute muscle injury and muscular dystrophy
 Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Material Levels of protein drive the reparative process in acute muscle injury and muscular dystrophy Francesca Riuzzi 1,4 *, Sara Beccafico 1,4 *, Roberta Sagheddu 1,4, Sara Chiappalupi
Supplementary Information
 Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
Supplementary Information promotes cancer cell invasion and proliferation by receptor-mediated endocytosis-dependent and -independent mechanisms, respectively Kensaku Shojima, Akira Sato, Hideaki Hanaki,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
Engineering Nanomedical Systems. Assessing the efficacy of nanomedical therapies
 BME 626 October 9, 2014 Engineering Nanomedical Systems Lecture 12 Assessing the efficacy of nanomedical therapies James F. Leary, Ph.D. SVM Endowed Professor of Nanomedicine Professor of Basic Medical
BME 626 October 9, 2014 Engineering Nanomedical Systems Lecture 12 Assessing the efficacy of nanomedical therapies James F. Leary, Ph.D. SVM Endowed Professor of Nanomedicine Professor of Basic Medical
efluor Organic Dyes 450/50 BP Fluorescence Intensity Wavelength (nm)
 efluor Organic Dyes efluor Organic Dyes Catalog No. Description Clone Application Anti-Mouse efluor 450 Products 48-0042 Anti-mouse CD4 RM4-5 Flow Cytometry 48-0081 Anti-mouse CD8 53-6.7 Flow Cytometry
efluor Organic Dyes efluor Organic Dyes Catalog No. Description Clone Application Anti-Mouse efluor 450 Products 48-0042 Anti-mouse CD4 RM4-5 Flow Cytometry 48-0081 Anti-mouse CD8 53-6.7 Flow Cytometry
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Fig. S1. Nature Medicine: doi: /nm HoxA9 expression levels BM MOZ-TIF2 AML BM. Sca-1-H. c-kit-h CSF1R-H CD16/32-H. Mac1-H.
 A 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 1 1 1 1 1 2 1 3 1 4 GFPH 1 1 1 1 1 2 1 3 1 4 Sca1H 1 1 1 1 1 2 1 3 1 4 ckith 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH
A 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 1 1 1 1 1 2 1 3 1 4 GFPH 1 1 1 1 1 2 1 3 1 4 Sca1H 1 1 1 1 1 2 1 3 1 4 ckith 1 4 1 4 1 4 CSF1RH 1 3 1 2 1 1 CSF1RH 1 3 1 2 1 1 CSF1RH
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs
 FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
Supporting Information
 Supporting Information Liu et al. 10.1073/pnas.0901216106 SI Materials and Methods Reagents. Thioglycolate and LPS from Escherichia coli 0111:B4 were from Sigma-Aldrich. PAM3CSK4 and poly(i:c) were from
Supporting Information Liu et al. 10.1073/pnas.0901216106 SI Materials and Methods Reagents. Thioglycolate and LPS from Escherichia coli 0111:B4 were from Sigma-Aldrich. PAM3CSK4 and poly(i:c) were from
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
of signal transduction pathways?
 CELL BIOLOGY Signal Transduction + How do you study activation of signal transduction pathways? + IDENTIFICATION OF PATHWAY-SPECIFIC TRANSCRIPTION ACTIVATION + EASIER, FASTER THAN BLOTTING OR GEL-SHIFT
CELL BIOLOGY Signal Transduction + How do you study activation of signal transduction pathways? + IDENTIFICATION OF PATHWAY-SPECIFIC TRANSCRIPTION ACTIVATION + EASIER, FASTER THAN BLOTTING OR GEL-SHIFT
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplementary information
 Supplementary information Inhibition of mitochondrial dysfunction and neuronal cell death in models of Huntington s disease and in HD patients-derived cells Xing Guo 1,#, Marie-Helene Disatnik 3,#, Marie
Supplementary information Inhibition of mitochondrial dysfunction and neuronal cell death in models of Huntington s disease and in HD patients-derived cells Xing Guo 1,#, Marie-Helene Disatnik 3,#, Marie
Supplementary Material. TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the. ERK1/2 signaling pathway
 Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
Supplementary Material TRIB3 inhibits proliferation and promotes osteogenesis in hbmscs by regulating the ERK1/2 signaling pathway Cui Zhang 1, Fan-Fan Hong 1, Cui-Cui Wang 1, Liang Li 1, Jian-Ling Chen
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
BD Cytometric Bead Array. Multiplexed Bead-Based Immunoassays
 BD Cytometric Bead Array Multiplexed Bead-Based Immunoassays Cover Illustration: Fairman Studios, LLC. BD Cytometric Bead Array Multiplexed Bead-Based Immunoassays BD Cytometric Bead Array (CBA) is a flow
BD Cytometric Bead Array Multiplexed Bead-Based Immunoassays Cover Illustration: Fairman Studios, LLC. BD Cytometric Bead Array Multiplexed Bead-Based Immunoassays BD Cytometric Bead Array (CBA) is a flow
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit
 RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
RayBio Human NF-κB p65 Transcription Factor Activity Assay Kit Catalog #: TFEH-p65 User Manual Mar 13, 2017 3607 Parkway Lane, Suite 200 Norcross, GA 30092 Tel: 1-888-494-8555 (Toll Free) or 770-729-2992,
Supplemental Information. Lithocholic Acid Hydroxyamide Destabilizes. Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting. Deubiquitinase USP2a
 Cell Chemical Biology, Volume 24 Supplemental Information Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting Deubiquitinase USP2a Katarzyna Magiera, Marcin Tomala,
Cell Chemical Biology, Volume 24 Supplemental Information Lithocholic Acid Hydroxyamide Destabilizes Cyclin D1 and Induces G 0 /G 1 Arrest by Inhibiting Deubiquitinase USP2a Katarzyna Magiera, Marcin Tomala,
Species predicted to react based on 100% sequence homology: Chicken, Bovine, Dog.
 1 of 5 11/1/2013 10:25 PM Product Pathways - Jak/Stat Pathway Phospho-Stat3 (Tyr705) Antibody #9131 Have you tried your application using our XP monoclonal antibodies? Try products: 9145 PhosphoSitePlus
1 of 5 11/1/2013 10:25 PM Product Pathways - Jak/Stat Pathway Phospho-Stat3 (Tyr705) Antibody #9131 Have you tried your application using our XP monoclonal antibodies? Try products: 9145 PhosphoSitePlus
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Supplemental Information. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis
 Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Regulation and Function of the IL-1 Family Cytokine IL-1F9 in Human Bronchial Epithelial Cells
 Online Data Supplement Regulation and Function of the IL-1 Family Cytokine IL-1F9 in Human Bronchial Epithelial Cells Regina T. Chustz 1, Deepti R. Nagarkar 1, Julie A. Poposki 1, Silvio Favoreto, Jr 1,
Online Data Supplement Regulation and Function of the IL-1 Family Cytokine IL-1F9 in Human Bronchial Epithelial Cells Regina T. Chustz 1, Deepti R. Nagarkar 1, Julie A. Poposki 1, Silvio Favoreto, Jr 1,
Supplemental Information. Loss of MicroRNA-7 Regulation Leads. to a-synuclein Accumulation and. Dopaminergic Neuronal Loss In Vivo
 YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
Supplementary Data, Romain et al., ATVB/2012/300516R2
 Supplementary Data, Romain et al., ATV/212/3516R2 Methods online Mice All experiments were conducted on male mice between 8 and 12 weeks of age. Mice with genetic overexpression of SOCS3 or cell-specific
Supplementary Data, Romain et al., ATV/212/3516R2 Methods online Mice All experiments were conducted on male mice between 8 and 12 weeks of age. Mice with genetic overexpression of SOCS3 or cell-specific
The Effect of cdna on Tumor Cells Growth on Nude Mice
 The Effect of cdna on Tumor Cells Growth on Nude Mice Yiming Ding Department of Electric and Computer Engineering Portland State University, OR Email: dym@cecs.pdx.edu This project is assigned in the class
The Effect of cdna on Tumor Cells Growth on Nude Mice Yiming Ding Department of Electric and Computer Engineering Portland State University, OR Email: dym@cecs.pdx.edu This project is assigned in the class
HISTORY AND BACKGROUND
 Integrated Technology Platform Protein Kinases for Drug Development in Oncology Christoph Sachsenmaier and Christoph Schäechtele ProQinase GmbH, Freiburg, Germany BioTechniques 33:S101-S106 (October 2002)
Integrated Technology Platform Protein Kinases for Drug Development in Oncology Christoph Sachsenmaier and Christoph Schäechtele ProQinase GmbH, Freiburg, Germany BioTechniques 33:S101-S106 (October 2002)
Hossain_Supplemental Figure 1
 Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
Hossain_Supplemental Figure 1 GFP-PACT GFP-PACT Motif I GFP-PACT Motif II A. MG132 (1µM) GFP Tubulin GFP-PACT Pericentrin GFP-PACT GFP-PACT Pericentrin Fig. S1. Expression and localization of Orc1 PACT
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Materials and Methods:
 Supplementary Materials and Methods: Preclinical chemoprevention experimental design Mice were weighed and each mammary tumor was manually palpated and measured with digital calipers once a week from 4
Supplementary Materials and Methods: Preclinical chemoprevention experimental design Mice were weighed and each mammary tumor was manually palpated and measured with digital calipers once a week from 4
Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using
 Supplementary Methods Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using the Qiagen RNeasy Mini Kit, according to the manufacturer s protocol with on-column DNase digestion
Supplementary Methods Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using the Qiagen RNeasy Mini Kit, according to the manufacturer s protocol with on-column DNase digestion
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer
 Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
Supplementary Figure 1 Characterization of sirna-onv stability. (a) Fluorescence recovery curves of SQ-siRNA-ONV and SQ-ds-siRNA in 1 TAMg buffer containing 10% serum The data error bars indicate means
BF Fox-3 HLA merge A 3
 Supplementary Figure 1 F Fox-3 merge 3 SN TRL L P L TRL SN P Supplementary Figure 1. Series of low magnification brightfield and immunofluorescence images (Fox-3 in green/, and in red). ircles indicate
Supplementary Figure 1 F Fox-3 merge 3 SN TRL L P L TRL SN P Supplementary Figure 1. Series of low magnification brightfield and immunofluorescence images (Fox-3 in green/, and in red). ircles indicate
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
TITLE: Novel Small-Molecule Inhibitor of Tyk2: Lucrative Therapeutic Target in Lupus
 AWARD NUMBER: W81XWH-16-1-0609 TITLE: Novel Small-Molecule Inhibitor of Tyk2: Lucrative Therapeutic Target in Lupus PRINCIPAL INVESTIGATOR: Abhishek Trigunaite CONTRACTING ORGANIZATION: SRI International
AWARD NUMBER: W81XWH-16-1-0609 TITLE: Novel Small-Molecule Inhibitor of Tyk2: Lucrative Therapeutic Target in Lupus PRINCIPAL INVESTIGATOR: Abhishek Trigunaite CONTRACTING ORGANIZATION: SRI International
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand
 SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
SUPPLEMENTAL FIGURES Beta3 integrin promotes long-lasting activation and polarization of Vascular Endothelial Growth Factor Receptor 2 by immobilized ligand C. Ravelli et al. FIGURE S. I Figure S. I: Gremlin
Supporting Information
 Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supporting Information Chakrabarty et al. 10.1073/pnas.1018001108 SI Materials and Methods Cell Lines. All cell lines were purchased from the American Type Culture Collection. Media and FBS were purchased
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
- NaCr. + NaCr. α H3K4me2 α H3K4me3 α H3K9me3 α H3K27me3 α H3K36me3 H3 H2A-2B H4 H3 H2A-2B H4 H3 H2A-2B H4. α Kcr. (rabbit) α Kac.
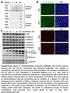 + NaCr NaCr + NaCr NaCr Peptides 10ng 50ng 250ng K α Pan (mouse) Pan (mouse) 10ng 50ng 250ng α Pan (rabbit) C 10ng 50ng 250ng α Pan (mouse) 0 1.25 2.5 5 10 20 40 (mm) NaCr 24h α Pan (rabbit) α K4me2 α
+ NaCr NaCr + NaCr NaCr Peptides 10ng 50ng 250ng K α Pan (mouse) Pan (mouse) 10ng 50ng 250ng α Pan (rabbit) C 10ng 50ng 250ng α Pan (mouse) 0 1.25 2.5 5 10 20 40 (mm) NaCr 24h α Pan (rabbit) α K4me2 α
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary information
 Supplementary information Cooperation of Oncolytic Herpes Virotherapy and PD-1 Blockade in Murine Rhabdomyosarcoma Models Chun-Yu Chen 1, Pin-Yi Wang 1, Brian Hutzen 1, Les Sprague 3, Hayley M. Swain 1,
Supplementary information Cooperation of Oncolytic Herpes Virotherapy and PD-1 Blockade in Murine Rhabdomyosarcoma Models Chun-Yu Chen 1, Pin-Yi Wang 1, Brian Hutzen 1, Les Sprague 3, Hayley M. Swain 1,
Therapeutic RNA delivery
 Therapeutic RNA delivery 05/02/2017 TIDES Dr. Adrien Weingärtner Head of Technology Development Forward looking statements THE INFORMATION CONTAINED IN THIS PRESENTATION IS BEING SUPPLIED AND COMMUNICATED
Therapeutic RNA delivery 05/02/2017 TIDES Dr. Adrien Weingärtner Head of Technology Development Forward looking statements THE INFORMATION CONTAINED IN THIS PRESENTATION IS BEING SUPPLIED AND COMMUNICATED
Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications
 PRODUCT FAMILY BULLETIN Tropix Chemiluminescent Kits and Reagents Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications Introduction to Chemiluminescence Chemiluminescence is the conversion
PRODUCT FAMILY BULLETIN Tropix Chemiluminescent Kits and Reagents Tropix Chemiluminescent Kits and Reagents For Cell Biology Applications Introduction to Chemiluminescence Chemiluminescence is the conversion
TheraLin. Universal Tissue Fixative Enabling Molecular Pathology
 TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
Supplemental Information. Inflammatory Ly6C high Monocytes Protect. against Candidiasis through IL-15-Driven. NK Cell/Neutrophil Activation
 Immunity, Volume 46 Supplemental Information Inflammatory Ly6C high Monocytes Protect against Candidiasis through IL-15-Driven NK Cell/Neutrophil Activation Jorge Domínguez-Andrés, Lidia Feo-Lucas, María
Immunity, Volume 46 Supplemental Information Inflammatory Ly6C high Monocytes Protect against Candidiasis through IL-15-Driven NK Cell/Neutrophil Activation Jorge Domínguez-Andrés, Lidia Feo-Lucas, María
Supplemental Materials and Methods
 Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
