Nature Biotechnology: doi: /nbt Supplementary Figure 1. Design and sequence of system 1 for targeted demethylation.
|
|
|
- Gordon Nicholas Scott
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1 Design and sequence of system 1 for targeted demethylation. (a) Design of system 1 for targeted demethylation. TET1CD was fused to a catalytic inactive Cas9 nuclease (dcas9) and expressed using the CAG promoter (pcag-dcas9tet1cd). Each grna vector was generated by incorporating the target sequence into the grna cloning vector with the U6 promoter (Addgene, 41824) using Gibson assembly (New England BioLabs). (b) Full sequence of pcag-dcas9tet1cd.
2 Supplementary Figure 2 Design and sequence of system 2 for targeted demethylation. (a) Design of system 2 for targeted demethylation. dcas9 with ten copies of the GCN4 peptide was expressed using the CAG promoter (pcag-dcas9-10xgcn4_v4). The length of the linker separating each 19 aa GCN4 peptide unit of the array was 5 aa. An anti-gcn4 peptide antibody (scfv)-sfgfp-tet1cd fusion protein was expressed using the CAG promoter (pcag-scfvgcn4sfgfptet1cd). Each grna vector was generated by incorporating the target sequence into the grna cloning vector with the U6 promoter (Addgene, 41824) using Gibson assembly (New England BioLabs). (b) Full sequence of pcag-dcas9-10xgcn4_v4. (c) Full sequence of scfvgcn4sfgfptet1cd.
3 Supplementary Figure 3 Design and sequence of system 3 for targeted demethylation. (a) Design of system 3 for targeted demethylation. dcas9 with five copies of the GCN4 peptide was expressed using the CAG promoter (pcag-dcas9-5xplat2afld). The length of the linker separating each 19 aa GCN4 peptide unit of the array was 22 aa. An anti-gcn4 peptide antibody (scfv)-sfgfp-tet1cd fusion protein was expressed using the CAG promoter (pcag-scfvgcn4sfgfptet1cd). Each grna vector was generated by incorporating the target sequence into the grna cloning vector with the U6 promoter (Addgene: 41824) using Gibson assembly (New England BioLabs). (b) Full sequence of pcag-dcas9-5xplat2afld.
4 Supplementary Figure 4 Design and sequence of system 4 for targeted demethylation. (a) Design of system 4 for targeted demethylation. dcas9 with four copies of the GCN4 peptide was expressed using the CAG promoter (pcag-dcas9-3.5xsuper). The length of the linker separating each 19 aa GCN4 peptide unit of the array was 43 aa. An anti-gcn4 peptide antibody (scfv)-sfgfp-tet1cd fusion protein was expressed using the CAG promoter (pcag-scfvgcn4sfgfptet1cd). Each grna vector was generated by incorporating the target sequence into the grna cloning vector with the U6 promoter (Addgene: 41824) using Gibson assembly (New England BioLabs). (b) Full sequence of pcag-dcas9-3.5xsuper.
5 Supplementary Figure 5 Validation of linker length and specificity of demethylation. (a) The length of the linker separating each GCN4 peptide unit of the array fused to dcas9 is important for maximization of the demethylation activity. There are two possibilities to explain this. If the linker is too short, there is only a small amount of space for antibody-tet1cd fusion proteins to approach and bind to the GCN4 peptide array, resulting in poor demethylation activity (possibility 1). Alternatively, the amount of space is too small for antibody-tet1cd fusion proteins to work properly, resulting in poor demethylation activity (possibility 2). (b) Co-IP of the Cas9-peptide array and antibody-sfgfp-tet1cd fusion protein (systems 2 4). The Cas9- peptide array (with a HA tag) was immunoprecipitated with anti-ha magnetic beads and subjected to western blot analysis. The amount of co-immunoprecipitated antibody-sfgfp-tet1cd fusion protein (vector lacking the HA tag was used in this case) was quantified by western blotting with an anti-gfp antibody and normalized by the amount of the Cas9-peptide array quantified by western blotting with an anti-ha antibody. Normalized values are shown in the bar graph. Data are shown as the mean ± s.e.m. Statistical analyses were performed using an ANOVA with Tukey s post-hoc test (N.S., not significant (p > 0.05)). (c) Demethylation activities of system 3 (dcas9-gcn4 and scfv-gfp-tet1cd), that with catalytic inactive TET1 (H1671Y, D1673A), that without scfv-gfp-tet1cd, and that without dcas9-gcn4. Demethylation of the STAT3-binding site was analyzed by COBRA. The grna used was target 2 of Gfap. Demethylation was analyzed as in Figure 1c. Data are shown as the mean ± s.e.m. (n = 3 from two independent experiments). The twosided Student s t-test was performed. ***P <
6 Supplementary Figure 6 Quantification of the viability of ESCs into which systems 2, 3, and 4 were introduced. (a) ESCs were transfected with the Gfap_2 grna using systems 2, 3, and 4. Two days after transfection, ESCs were stained with PI, and cell viability was quantified using a FACSVerse flow cytometer (BD Biosciences) with a 488-nm blue laser. Cells were categorized into four groups based on dye uptake and GFP signals. Quadrant UL shows PI-negative/GFP-positive cells, quadrant UR shows PI/GFP-positive cells, quadrant LL shows PI/GFP-negative cells, and quadrant LR shows PI-positive/GFP-negative cells. (b) The population of PI-positive cells among GFP-positive cells was compared among the systems. Data are shown as the mean ± s.e.m (n = 3 from two independent experiments). Statistical analyses were performed using an ANOVA with Tukey s post-hoc test (N.S., not significant (p > 0.05)).
7 Supplementary Figure 7 Design and sequence of the all-in-one system for targeted demethylation. (a) Design of the all-in-one system for targeted demethylation. This vector included the grna under the control of the U6 promoter, dcas9 with the GCN4 array (system 3), and the antibody-tet1cd fusion protein. Cloning was performed by linearization of an Afl II site and Gibson assembly-mediated incorporation of the grna insert fragment. (b) Full sequence of pplattet-grna2.
8 Supplementary Figure 8 Methylation surrounding the target site. ESCs transfected with Gfap grna (Gfap_2) or a control using systems 2, 3 (same as Figure 1f), and 4 were sorted to isolate GFPexpressing cells, and methylation in the surrounding area was analyzed using bisulfite sequencing. Methylation for active and catalytically-dead TET1 is shown. Black/white circles indicate the percentage of methylation in each CpG site. Black indicates the methylation percentage. Each number beneath the circles indicates the position. The red bar indicates the STAT3-binding site. The blue bar indicates the target site. A scale is provided at the bottom. For each group, at least 14 randomly selected clones were sequenced and analyzed. The statistical significance between the two groups of the entire set of CpG sites was evaluated with the Mann-Whitney U-test (also called the Wilcoxon rank-sum test).
9 Supplementary Figure 9 Off-target analysis and genome-wide analysis of DNA methylation and gene expression. (a) Methylation surrounding the off-target sites of the Gfap_2 grna. ESCs were transfected with this grna or a control using system 3, sorted, and analyzed for methylation in the area surrounding off-target sites by bisulfite sequencing. Matched sequences are written in red and mismatched sequences are written in black. Red bars indicate the homologous sequence to the Gfap target sites. Black/white circles indicate the percentage of methylation in each CpG site. Black indicates the methylation percentage. Each number beneath the circles indicates the position. A scale is provided at the bottom. For each group, at least 14 randomly selected clones were sequenced and analyzed. The statistical significance between the two groups of the entire set of CpG sites was evaluated with the Mann-Whitney U-test (also called the Wilcoxon rank-sum test). (b) BS-seq analysis of ESCs that were transfected with the all-in-one vector targeting Gfap (samples 1 and 3) or control (samples 2 and 4). Vectors with active (samples 1 and 2) or catalytically-dead (samples 3 and 4) TET1 were used. Sample 1 and 2 was mixed and randomly divided in half to generate samples a and b. Scatter plots of 1 vs. 2, 3 vs. 4, 1 vs. 3, and a vs. b are shown, along with the correlation coefficients. Each red line indicates the regression line of each sample. (c) BS-seq methylation landscape of the Gfap gene in samples 1 4. The STAT3-binding site and corresponding methylation are indicated by a pink bar and a pink square, respectively. (d) RNA-seq analysis of the same samples as in Supplementary Figure 9b. Scatter plots of 1 vs. 2, 3 vs. 4, and 1 vs. 3 are shown, along with the correlation coefficients.
10 Supplementary Figure 10 Methylation surrounding the H19 DMR CTCF-binding sites (m1 and m2). ESCs were transfected with H19 DMR_2 grna using system 3, sorted, and analyzed for methylation in the area surrounding target site 2 by bisulfite sequencing. Methylation for active and catalytically-dead TET1 is shown. Black/white circles indicate the percentage of methylation in each CpG site. Black indicates the methylation percentage. Each number beneath the circles indicates the position. Red bars indicate the CTCF-binding sites (m1 and m2). The blue bar indicates the position of target 2. A scale is provided at the bottom. For each group, at least 14 randomly selected clones were sequenced and analyzed. The statistical significance between the two groups of the entire set of CpG sites was evaluated with the Mann-Whitney U-test (also called the Wilcoxon rank-sum test).
11 Supplementary Figure 11 Methylation surrounding the off-target sites of the H19DMR 1 4 grnas. ESCs were transfected with these grnas using system 3, sorted, and analyzed for methylation in the area surrounding off-target sites by bisulfite sequencing. Matched sequences are written in red and mismatched sequences are written in black. Red bars indicate the homologous sequence to the H19 target sites. Black/white circles indicate the percentage of methylation in each CpG site. Black indicates the methylation percentage. Each number beneath the circles indicates the position. A scale is provided at the bottom. For each group, at least 14 randomly selected clones were sequenced and analyzed. The statistical significance between the two groups of the entire set of CpG sites was evaluated with the Mann-Whitney U-test (also called the Wilcoxon rank-sum test).
12 Supplementary Figure 12 Expression and methylation analysis of methylation-edited cells. (a) Expression analysis of methylation-edited cells. Cells were transfected with grnas for RHOXF2B, CARD9, SH3BP2, or CNKSR1 using system 3 with active or catalytically-dead TET1CD. The cells were sorted and analyzed for expression by quantitative PCR. Data are shown as the mean ± s.e.m. (n = 3 from two independent experiments). The two-sided Student s t-test was performed. N.S., not significant; **P < 0.01; ***P < (b) Demethylation of the RHOXF2B, CARD9, SH3BP2, and CNKSR1 genes in human cells using system 3 with sorting. Demethylation activities for active and catalytically-dead TET1 are shown. Demethylation was analyzed as in Figure 1c. Data are shown as the mean ± s.e.m. (n = 3 from two independent experiments). The two-sided Student s t-test was performed. N.S., not significant; ***P <
13 Supplementary Figure 13 E18 brain sections that were electroporated with the vector targeting Gfap or the control vector at E14. (a) E18 brain sections that were electroporated with the vector targeting Gfap or the control vector at E14. Brain sections obtained using catalytically-dead TET1 are also shown. Green, red, and blue indicate GFP, GFAP, and Hoechst, respectively. Magnified images of the boxed areas indicated are also shown in 1, 2, 1, and 2, respectively. The population of GFAP-positive cells among GFP-positive cells was significantly increased compared to the control. A scale bar is provided in the image. (b) The percentage of GFAP-positive cells among GFP-positive cells. The results obtained using catalytically-dead TET1 are also shown. Cortical sections at the same anatomical level were analyzed, and confocal images were taken with a confocal microscope. To assess astrocyte differentiation, at least 300 GFP-positive cells per sample (n=4 6 brains per group) were counted. GFAP-positive cells among GFP-positive cells were counted in high-magnification images, and each GFAP-positive cell was identified by GFAP staining around the nucleus, as indicated by both GFP and Hoechst. Data are shown as the mean ± s.e.m. Statistical analyses were performed using an ANOVA with Tukey s post-hoc test (*p < 0.05, **p < 0.01, ***p < 0.001). (c) DNA methylation of the Gfap locus in GFP-positive cells in fetal brains that were electroporated with the vector targeting Gfap (Gfap_2, Gfap_3, and Gfap_0) or the control vector. The results obtained with catalytically-dead TET1 are also shown. GFP-positive cells were electroporated with the vectors at E14. At 24 h after electroporation, GFP-positive cells were sorted by FACS and used for DNA methylation analysis. The average methylation of CpGs at the Gfap locus analyzed by bisulfite sequencing is presented. At least three samples were used for analysis. Data are shown as the mean ± s.e.m. Statistical analyses were performed using an ANOVA with Tukey's post-hoc test (*p < 0.05).
14 Supplementary Figure 14 Luciferase reporter assay of the Gfap promoter. A neural progenitor cell line from adult rat hippocampus (HCN cells) was co-transfected with the all-in-one vector (expressing grna for the control or Gfap locus) and a Gfap promoter-reporter plasmid, which expresses firefly luciferase under the regulation of the 2.6 kb Gfap promoter. The reporter assay using catalytically-dead TET1 is also shown. As an internal control, the sea pansy luciferaseexpressing vector under the control of the human elongation factor-1a promoter was also co-transfected. One day after transfection, cells were stimulated with LIF (50 ng/ml) for 8 h and used for luciferase analysis. Firefly luciferase activities were determined by three independent transfections and normalized by comparison with Renilla luciferase activities as the internal control. Data are shown as the mean ± s.e.m. Statistical analyses were performed using an ANOVA with Tukey s post-hoc test (*p < 0.05). ns, not significant (p > 0.05).
15 Supplementary Figure 15 A flowchart of the selection of off-target sites for bisulfite sequencing analysis. Off-targets sites were searched for using a web tool called CRISPR direct ( By using this tool, the 12 bases in the 3 region of the target sequence adjacent to the PAM were searched against the genome because this region contains critical residues determining target specificity. Next, the sites unsuitable for analysis (sequences containing repeats and those giving no PCR primers by Meth Primer ( in the default condition except for the product size) were removed. As for the offtarget analysis of Gfap, all these sites were selected and subjected to off-target analysis. As for the off-targets of H19, at least all the off-targets in which more than 16 of 20 bases match were selected. If there were fewer than three selected sites, sites of lower homology were selected.
16 Target sequences Target name Target sequence Methylation-sensitive site near the targets Gfap_0 GTGAGACCACCTTCTGCCTCTGG Gfap STAT3-binding site Gfap_1 ATAGACATAATGGTCAGGGGTGG Gfap STAT3-binding site Gfap_2 GGATGCCAGGATGTCAGCCCCGG Gfap STAT3-binding site Gfap_3 ATATGGCAAGGGCAGCCCCGTGG Gfap STAT3-binding site H19DMR_1 GTGGGGGGGCTCTTTAGGTTTGG H19 DMR CTCF-binding site 1 H19DMR_2 ACCCTGGTCTTTACACACAAAGG H19 DMR CTCF-binding site 2 H19DMR_3 GAAGCTGTTATGTGCAACAAGGG H19 DMR CTCF-binding site 3 H19DMR_4 CAGATTTGGCTATAGCTAAATGG H19 DMR CTCF-binding site 4 RHOXF2B GCTTGGCCTTGCCCGGATGAGGG RHOXF2B promoter CARD9 TGGGAGCAGCTTTCCTCTGGAGG CARD9 promoter SH3BP2 TGAGGTCCTGAAAGCTGCCTGGG SH3BP2 promoter CNKSR1 TGTGAGCCCAGGTATGCAGTAGG CNKSR1 promoter PAM sequences are indicated in red. Sequences of unrelated grnas Target name UR_1 UR_2 UR_3 grna sequence CCATTATTGCATTAATCTGA TAATGCAGCCAGAAAATGAC TCAGGGATCAAATTCTGAGC
17 COBRA primer sequences Primer name Primer sequence Restriction enzyme Methylation-sensitive site near the targets GfapSTAT3-B1 GTTGAAGATTTGGTAGTGTTGAGTT Hpy188III Gfap STAT3-binding site GfapSTAT3-B2 TAAAACATATAACAAAAACAACCCC H19DMR-B1 AAGGAGATTATGTTTTATTTTTGGA BstUI H19 DMR CTCF-binding site 1 H19DMR-B2 AAAAAAACTCAATCAATTACAATCC H19DMR-B1 AAGGAGATTATGTTTTATTTTTGGA RsaI H19 DMR CTCF-binding site 2 H19DMR-B2 AAAAAAACTCAATCAATTACAATCC H19DMR-B3 GGGTTTTTTTGGTTATTGAATTTTAA BstUI H19 DMR CTCF-binding site 3 H19DMR-B4 AATACACACATCTTACCACCCCTATA H19DMR-B5 TTTTTGGGTAGTTTTTTTAGTTTTG BstUI H19 DMR CTCF-binding site 4 H19DMR-B6 ACACAAATACCTAATCCCTTTATTAAAC RHOXF2B-B1 GTTATAAAATGGGTTTGTATAATTTAGTAT BstUI RHOXF2B promoter RHOXF2B-B2 AAAACCTCCTCTCTTACTTTTCTACTTC CARD9-B3 GGTTATTAGGGATTGTTTTTTTGTG Aci I CARD9 promoter CARD9-B4 ATCTTCCAAAAACCACCTACACTAC SH3BP2-B1 TTATAGGGTAGAAGGTAGGAAGTGT Aci I SH3BP2 promoter SH3BP2-B2 ATCTCCCAAACATATAAAACCTAAC CNKSR1-B1 TTTTTTTAGGTTTGGGTTTTGG Taq I CNKSR1 promoter CNKSR1-B2 AATAACCCACCCACCTTAACTTC
18 Bisulfite sequencing PCR primer sequences Primer name Primer sequence Methylation-sensitive site near the targets GfapSTAT3-B3 TTGGTTAGTTTTTAGGATTTTTTTT Gfap STAT3-binding site (ES) GfapSTAT3-B4 AAAACTTCAAACCCATCTATCTCTTC GFmS GGGATTTATTAGGAGAATTTTAGTAAGTAG Gfap STAT3-binding site (primary culture) GFmAS TCTACCCATACTTAAACTTCTAA TATCTAC H19DMR-B1 AAGGAGATTATGTTTTATTTTTGGA H19 DMR CTCF-binding site 1 H19DMR-B2 AAAAAAACTCAATCAATTACAATCC Gfap_O1B1 TTGTAAAGGTAGGATTAATAAGGGAATT Gfap off-target site 1 Gfap_O1B2 AAAAAAAACCCTTCAAAAAAAATCTA Gfap_O2B1 TTATTATTTATATTTGGAGGGAGGG Gfap off-target site 2 Gfap_O2B2 ATTACACCAAAAAAATTTTAAAAAC Gfap_O3B1 TTTAAATTTTTTTATGTGAATATGG Gfap off-target site 3 Gfap_O3B2 AAACATTTAATTCATTAATACACAC Gfap_O4B1 TTTTAAGTTTTTAGGATGAGAAAGA Gfap off-target site 4 Gfap_O4B2 AAAATTATTTCCAATAAACTACCCC Gfap_O5B1 ATTATTTTTGTGGATTGTTTTAGGG Gfap off-target site 5 Gfap_O5B2 AACCACCAAAAATTACATAAACTCC Gfap_O6B1 TGGGAGAAGTTTTTAGGAGTATGAG Gfap off-target site 6 Gfap_O6B2 ACAAATAAAAAAACCACAAAAAAACA Gfap_O7B1 TTAGTTTGGAATTTTGAGGTTAGTAGTTT Gfap off-target site 7 Gfap_O7B2 AAACATCTTACAATACAATAAACATTTACA Gfap_O8B1 TGTTTATTTTAAGGTAATAAAGATTTAGT Gfap off-target site 8 Gfap_O8B2 ATAAAATTATCAAATCTCCATATATTACTT Gfap_O9B1 TGTTGTTGTAAGTTAGGGGTAGGTT Gfap off-target site 9 Gfap_O9B2 ATTTTCCCACCACACTAAAAATTAC Gfap_O10B1 TTGTATTTTTTTAGGGTTGTTTTTTAATT Gfap off-target site 10 Gfap_O10B2 CACACATACCAAAATATACCAATCAC H19DMR1OT1-B1 GAGGGTATTGTATATTTGAAGGAGTTT H19DMR1 off-target site 1 H19DMR1OT1-B2 AATTCTAAAAAACAACAAACTTATCTCACT H19DMR1OT2-B1 TTTTTTTTGTTAAAGGTAAGGAAA H19DMR1 off-target site 2 H19DMR1OT2-B2 CAACAAAACCATCATAACCTACAAA H19DMR1OT3-B1 TGTTATTTTTGAGTTTTAATAGTTTTAGAA H19DMR1 off-target site 3 H19DMR1OT3-B2 ATAAACCCCAACCTAAAAAACAAAC H19DMR2OT1-B1 TTTTTTTATTATGTGGTTTAAGTT H19DMR2 off-target site 1 H19DMR2OT1-B2 ATACAAATTCAAAAAACATTTC H19DMR2OT2-B1 GATGTTATTGTTTGTTTTTTAAGAT H19DMR2 off-target site 2 H19DMR2OT2-B2 TTTCCAAAATTAATTTAAACCTC H19DMR2OT3-B1 TTAATAGATTATTTTTTGAATTAAAT H19DMR2 off-target site 3 H19DMR2OT3-B2 CTAATCACTATACTATATCTCACTAAAATT H19DMR3OT1-B1 GGATATTATTATTAATGTTTTAAGTATAA H19DMR3 off-target site 1 H19DMR3OT1-B2 CATTATATTACTTATCTATTTCCCC H19DMR3OT2-B1 TATTAAAGTTTAGTTGGTTTTTTTT H19DMR3 off-target site 2 H19DMR3OT2-B2 ACACACCATTATTACACATACTAAA H19DMR3OT3-B1 TTTGTTTTAGGAATTTTTGTGAAAAAT H19DMR3 off-target site 3 H19DMR3OT3-B2 TAATACCAAACAACCAAAATATCCC H19DMR4OT1-B1 AAATTAGTTTTATTTTGTTTTAAGTTT H19DMR3 off-target site 1 H19DMR4OT1-B2 TTATATTAAAATTCAATACTCTTAACAATT H19DMR4OT2-B1 ATAGGGTTTGGGATGAAATATTATG H19DMR3 off-target site 2 H19DMR4OT2-B2 ATCTCATTACTACTCACACATCAAAA
19 qpcr primer sequences Primer name Primer sequence Analysis GfapEx1S-1 GGAGAGGGACAACTTTGCAC Gfap expression analysis GfapEx2AS-1 ATACGCAGCCAGGTTGTTCT RHOXF2B-3 GGCAAGAAGCATGAATGTGA RHOXF2B expression analysis RHOXF2B-4 TGTCTCCTCCATTTGGCTCT H19Ex4,5-S1 TACCTGCCTCAGGAATCTGC H19 expression analysis H19Ex4,5-AS1 GTTGGCCATGAAGATGGATT Mus18S-S1 CCCGAAGCGTTTACTTTGAA Normalization for expression in ESCs Mus18S-AS1 CCCTCTTAATCATGGCCTCA ACTB sense GATGCAGAAGGAGATCACTGC Normalization for expression in HEK293 ACTB antisense GTACTTGCGCTCAGGAGGAG CARD9-1 CAGGCTCCTGGTGTGTCTG CARD9 expression analysis CARD9-2 CTCCAGCACTCGTCATCGT SH3BP2-1 ATGTGTTGGGTCAGCACCA SH3BP2 expression analysis SH3BP2-2 CAGGCATGGTTAGCAGGTTC CNKSR1-1 GGCAAAACAGGAGCTGATTC CNKSR1 expression analysis CNKSR1-2 TAGTCCTGCAGGGAGTCGTC
20 Gfap off-targets Total number: 20 Analyzable site for BS-seq Name chr start end Target homology sequence (red indicates matched sequence) Matched bases Homology (%) GTGACACAGGATGTCAGCCCGGG CCATGCTGGGATGTCAGCCCTGG GTCACCTTGGATGTCAGCCCCGG CCAGGTCAGGATGTCAGCCCAGG AGAAGGAAGGATGTCAGCCCAGG TGGAGCCTGGATGTCAGCCCAGG TATTGTCAGGATGTCAGCCCTGG ATTTTTCTGGATGTCAGCCCTGG ACACCAAAGGATGTCAGCCCAGG TTATGAGTGGATGTCAGCCCAGG 15 75
21 H19DMR1 off-targets Total number: 10 Selected for BS-seq Name chr start end Target homology sequence (red indicates matched sequence) Matched bases Homology (%) H19DMR1OT1 chr CTGTGCCAGCTCTTTAGGTTTGG H19DMR1OT2 chr CTTGATTTGCTCTTTAGGTTGGG H19DMR1OT3 chr AAAAGCCTGCTCTTTAGGTTGGG H19DMR2 off-targets Total number: 23 Selected for BS-seq Name chr start end Target homology sequence (red indicates matched sequence) Matched bases Homology (%) H19DMR2OT1 chr AAACAAGCCTTTACACACAAAGG H19DMR2OT2 chr GCCAAGGACTTTACACACAAGGG H19DMR2OT3 chr ACAGTTAGCTTTACACACAAAGG H19DMR3 off-targets Total number: 19 Selected for BS-seq Name chr start end Target homology sequence (red indicates matched sequence) Matched bases Homology (%) H19DMR3OT1 chr AAATCTGTTATGTGCAACAAGGG H19DMR3OT2 chr TATCCCAGTATGTGCAACAATGG H19DMR3OT3 chr ATCATCATTATGTGCAACAAGGG H19DMR4 off-targets Total number: 15 Selected for BS-seq No primers were found suitable to be designed on other off-targets. Name chr start end Target homology sequence (red indicates matched sequence) Matched bases Homology (%) H19DMR4OT1 chr CTGATGAAGCTATAGCTAAAGGG H19DMR4OT2 chr GGTTGTGAGCTATAGCTAAAGGG 13 65
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX
 Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Transfection of CRISPR/Cas9 Nuclease NLS ribonucleoprotein (RNP) into adherent mammalian cells using Lipofectamine RNAiMAX INTRODUCTION The CRISPR/Cas genome editing system consists of a single guide RNA
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Translation of HTT mrna with expanded CAG repeats is regulated by
 Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
Supplementary Figure 1 PCR-genotyping of the three mouse models used in this study and controls for behavioral experiments after semi-chronic Pten inhibition. a-c. DNA from App/Psen1 (a), Pten tg (b) and
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm
 Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Improving CRISPR-Cas9 Gene Knockout with a Validated Guide RNA Algorithm Anja Smith Director R&D Dharmacon, part of GE Healthcare Imagination at work crrna:tracrrna program Cas9 nuclease Active crrna is
Orflo Application Brief 3 /2017 GFP Transfection Efficiency Monitoring with Orflo s Moxi GO Next Generation Flow Cytometer. Introduction/Background
 Introduction/Background Cell transfection and transduction refer to an array of techniques used to introduce foreign genetic material, or cloning vectors, into cell genomes. The application of these methods
Introduction/Background Cell transfection and transduction refer to an array of techniques used to introduce foreign genetic material, or cloning vectors, into cell genomes. The application of these methods
RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors
 Supplementary Information RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors Pablo Perez-Pinera 1, Daniel D. Kocak 1, Christopher M. Vockley 2,3, Andrew F. Adler 1, Ami M. Kabadi 1,
Supplementary Information RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors Pablo Perez-Pinera 1, Daniel D. Kocak 1, Christopher M. Vockley 2,3, Andrew F. Adler 1, Ami M. Kabadi 1,
Chemical hijacking of auxin signaling with an engineered auxin-tir1
 1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
1 SUPPLEMENTARY INFORMATION Chemical hijacking of auxin signaling with an engineered auxin-tir1 pair Naoyuki Uchida 1,2*, Koji Takahashi 2*, Rie Iwasaki 1, Ryotaro Yamada 2, Masahiko Yoshimura 2, Takaho
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
 Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Xi et al. Genome Biology (2015) 16:231 DOI 10.1186/s13059-015-0791-1 RESEARCH A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132
 Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Neuron, Volume 65 Regulation of Synaptic Structure and Function by FMRP- Associated MicroRNAs mir-125b and mir-132 Dieter Edbauer, Joel R. Neilson, Kelly A. Foster, Chi-Fong Wang, Daniel P. Seeburg, Matthew
Genome editing. Knock-ins
 Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Genome editing Knock-ins Experiment design? Should we even do it? In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in
Nature Methods: doi: /nmeth.4396
 Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1 Comparison of technical replicate consistency between and across the standard ATAC-seq method, DNase-seq, and Omni-ATAC. (a) Heatmap-based representation of ATAC-seq quality control
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Nature Genetics: doi: /ng Supplementary Figure 1. Analysis of DHS STARR-seq in the P5424 cell line.
 Supplementary Figure 1 Analysis of DHS STARR-seq in the P5424 cell line. (a) Luciferase enhancer assays of proximal DHSs defined as active or inactive enhancers by STARR-seq in P5424 cells. For each candidate,
Supplementary Figure 1 Analysis of DHS STARR-seq in the P5424 cell line. (a) Luciferase enhancer assays of proximal DHSs defined as active or inactive enhancers by STARR-seq in P5424 cells. For each candidate,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
DOI: 10.1038/ncb2239 Hepatocytes (Endoderm) Foreskin fibroblasts (Mesoderm) Melanocytes (Ectoderm) +Dox rtta TetO CMV O,S,M,K & N H1 hes H7 hes H9 hes H1 hes-derived EBs H7 hes-derived EBs H9 hes-derived
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Multiplex Fluorescence Assays for Adherence Cells without Trypsinization
 Multiplex Fluorescence Assays for Adherence Cells without Trypsinization The combination of a bright field and three fluorescent channels allows the Celigo to perform many multiplexed assays. A gating
Multiplex Fluorescence Assays for Adherence Cells without Trypsinization The combination of a bright field and three fluorescent channels allows the Celigo to perform many multiplexed assays. A gating
Technical Bulletin. Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer. Introduction
 March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
March 212 Multiple Methods for Detecting Apoptosis on the BD Accuri C6 Flow Cytometer Contents 1 Introduction 2 Annexin V 4 JC-1 5 Caspase-3 6 APO-BrdU and APO-Direct Introduction Apoptosis (programmed
Direct Cell Counting Assays for Immuno Therapy
 Direct Cell Counting Assays for Immuno Therapy Cytotoxicity assays play a central role in studying the function of immune effector cells such as cytolytic T lymphocytes (CTL) and natural killer (NK) cells.
Direct Cell Counting Assays for Immuno Therapy Cytotoxicity assays play a central role in studying the function of immune effector cells such as cytolytic T lymphocytes (CTL) and natural killer (NK) cells.
Applications of Cas9 nickases for genome engineering
 application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
application note genome editing Applications of Cas9 nickases for genome engineering Shuqi Yan, Mollie Schubert, Maureen Young, Brian Wang Integrated DNA Technologies, 17 Commercial Park, Coralville, IA,
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
Supplemental Materials and Methods
 Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
Supplemental Materials and Methods 125 I-CXCL12 binding assay KG1 cells (2 10 6 ) were preincubated on ice with cold CXCL12 (1.6µg/mL corresponding to 200nM), CXCL11 (1.66µg/mL corresponding to 200nM),
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3'
 Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
Supplementary Table 1: Real-time PCR primer sequences for ΔEGFR, wtegfr, IL-6, LIF and GAPDH. Gene Sequence Fragment size ΔEGFR F 5' GGGCTCTGGAGGAAAAGAAAG GT 3' 116 bp R 5' CTTCTTACACTTGCGGACGC 3' wtegfr
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs
 FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
FIG S1: Calibration curves of standards for HPLC detection of Dopachrome (Dopac), Dopamine (DA) and Homovanillic acid (HVA), showing area of peak vs amount of standard analysed. Each point represents the
Chemically defined conditions for human ipsc derivation and culture
 Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Nature Methods Chemically defined conditions for human ipsc derivation and culture Guokai Chen, Daniel R Gulbranson, Zhonggang Hou, Jennifer M Bolin, Victor Ruotti, Mitchell D Probasco, Kimberly Smuga-Otto,
Application Note: Generating GFP-Tagged Human CD81 Tetraspanin Protein Using SBI s PrecisionX SmartNuclease System And HR Tagging Vectors
 Application Note: Generating GFP-Tagged Human CD81 Tetraspanin Protein Using SBI s PrecisionX SmartNuclease System And HR Tagging Vectors Table of Contents: I. Background Pg. 1 II. Analysis of Gene Target
Application Note: Generating GFP-Tagged Human CD81 Tetraspanin Protein Using SBI s PrecisionX SmartNuclease System And HR Tagging Vectors Table of Contents: I. Background Pg. 1 II. Analysis of Gene Target
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
EECS730: Introduction to Bioinformatics
 EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
EECS730: Introduction to Bioinformatics Lecture 14: Microarray Some slides were adapted from Dr. Luke Huan (University of Kansas), Dr. Shaojie Zhang (University of Central Florida), and Dr. Dong Xu and
Supplementary Information
 Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Supplementary Information MLL histone methylases regulate expression of HDLR- in presence of estrogen and control plasma cholesterol in vivo Khairul I. Ansari 1, Sahba Kasiri 1, Imran Hussain 1, Samara
Optimized, chemically-modified crrna:tracrrna complexes for CRISPR gene editing
 Optimized, chemically-modified crrna:tracrrna complexes for CRISPR gene editing Mark Behlke MD, PhD Chief Scientific Officer February 24, 2016 1 Implementing CRISPR/Cas9 gene editing 2 To focus on RNA
Optimized, chemically-modified crrna:tracrrna complexes for CRISPR gene editing Mark Behlke MD, PhD Chief Scientific Officer February 24, 2016 1 Implementing CRISPR/Cas9 gene editing 2 To focus on RNA
Cambridge University Press
 Figure 1.1. Model of RNAi pathway in C. elegans. Transmembrane protein SID-1 allows dsrna to enter the cell. In the cytoplasm,dsrna gets processed by DCR-1,existing in a complex with RDE-4,RDE-1 and DRH-1.
Figure 1.1. Model of RNAi pathway in C. elegans. Transmembrane protein SID-1 allows dsrna to enter the cell. In the cytoplasm,dsrna gets processed by DCR-1,existing in a complex with RDE-4,RDE-1 and DRH-1.
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest
 Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest C A. site-directed mutagenesis A C A T A DNA B. in vitro mutagenesis by PCR T A 1. anneal primer 1 C A 1. fill in
Lectures 28 and 29 applications of recombinant technology I. Manipulate gene of interest C A. site-directed mutagenesis A C A T A DNA B. in vitro mutagenesis by PCR T A 1. anneal primer 1 C A 1. fill in
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System
 ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
ChampionChIP Quick, High Throughput Chromatin Immunoprecipitation Assay System Liyan Pang, Ph.D. Application Scientist 1 Topics to be Covered Introduction What is ChIP-qPCR? Challenges Facing Biological
SUPPLEMENTARY MATERIAL AND METHODS
 SUPPLEMENTARY MATERIAL AND METHODS Amplification of HEV ORF1, ORF2 and ORF3 genome regions Total RNA was extracted from 200 µl EDTA plasma using Cobas AmpliPrep total nucleic acid isolation kit (Roche,
SUPPLEMENTARY MATERIAL AND METHODS Amplification of HEV ORF1, ORF2 and ORF3 genome regions Total RNA was extracted from 200 µl EDTA plasma using Cobas AmpliPrep total nucleic acid isolation kit (Roche,
Molecular Cloning. Joseph Sambrook. David W. Russell A LABORATORY MANUAL COLD SPRING HARBOR LABORATORY PRESS VOLUME.
 VOLUME Molecular Cloning A LABORATORY MANUAL THIRD EDITION www.molecularcloning.com Joseph Sambrook PETER MACCALLUM CANCER INSTITUTE AND THE UNIVERSITY OF MELBOURNE, AUSTRALIA David W. Russell UNIVERSITY
VOLUME Molecular Cloning A LABORATORY MANUAL THIRD EDITION www.molecularcloning.com Joseph Sambrook PETER MACCALLUM CANCER INSTITUTE AND THE UNIVERSITY OF MELBOURNE, AUSTRALIA David W. Russell UNIVERSITY
Supplementary Figure 1
 number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
Systematic Analysis of single cells by PCR
 Systematic Analysis of single cells by PCR Wolfgang Mann 27th March 2007, Weihenstephan Overview AmpliGrid Technology Examples DNA Forensics Single Cell Sequencing Polar Body Diagnostics Single Cell RT
Systematic Analysis of single cells by PCR Wolfgang Mann 27th March 2007, Weihenstephan Overview AmpliGrid Technology Examples DNA Forensics Single Cell Sequencing Polar Body Diagnostics Single Cell RT
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Protocol for tissue-specific gene disruption in zebrafish
 Protocol for tissue-specific gene disruption in zebrafish Overview This protocol describes a method to inactivate genes in zebrafish in a tissue-specific manner. It can be used to analyze mosaic loss-of-function
Protocol for tissue-specific gene disruption in zebrafish Overview This protocol describes a method to inactivate genes in zebrafish in a tissue-specific manner. It can be used to analyze mosaic loss-of-function
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Supplemental Information. Loss of MicroRNA-7 Regulation Leads. to a-synuclein Accumulation and. Dopaminergic Neuronal Loss In Vivo
 YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
YMTHE, Volume 25 Supplemental Information Loss of MicroRNA-7 Regulation Leads to a-synuclein Accumulation and Dopaminergic Neuronal Loss In Vivo Kirsty J. McMillan, Tracey K. Murray, Nora Bengoa-Vergniory,
A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging
 Resource A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging Marvin E. Tanenbaum, 1,2 Luke A. Gilbert, 1,2,3,4 Lei S. Qi, 1,3,4 Jonathan S. Weissman, 1,2,3,4 and
Resource A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging Marvin E. Tanenbaum, 1,2 Luke A. Gilbert, 1,2,3,4 Lei S. Qi, 1,3,4 Jonathan S. Weissman, 1,2,3,4 and
Alt-R CRISPR-Cpf1 System:
 user guide Alt-R CRISPR-Cpf1 System: Delivery of ribonucleoprotein complexes in HEK-293 cells using the Amaxa Nucleofector System See what more we can do for you at www.idtdna.com. For Research Use Only
user guide Alt-R CRISPR-Cpf1 System: Delivery of ribonucleoprotein complexes in HEK-293 cells using the Amaxa Nucleofector System See what more we can do for you at www.idtdna.com. For Research Use Only
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Quick reference guide
 Quick reference guide Our Invitrogen GeneArt CRISPR Search and Design Tool allows you to search our database of >600,000 predesigned CRISPR guide RNA (grna) sequences or analyze your sequence of interest
Quick reference guide Our Invitrogen GeneArt CRISPR Search and Design Tool allows you to search our database of >600,000 predesigned CRISPR guide RNA (grna) sequences or analyze your sequence of interest
CRISPR/Cas9 Genome Editing: Transfection Methods
 CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
Getting started with CRISPR: A review of gene knockout and homology-directed repair
 Getting started with CRISPR: A review of gene knockout and homology-directed repair Justin Barr Product Manager, Functional Genomics Integrated DNA Technologies 1 Agenda: Getting started with CRISPR Background
Getting started with CRISPR: A review of gene knockout and homology-directed repair Justin Barr Product Manager, Functional Genomics Integrated DNA Technologies 1 Agenda: Getting started with CRISPR Background
X2-C/X1-Y X2-C/VCAM-Y. FRET efficiency. Ratio YFP/CFP
 FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
Cold Fusion Cloning Kit. Cat. #s MC100A-1, MC101A-1. User Manual
 Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary figure 1 Flow diagram summarizing the overall experimental
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary figure 1 Flow diagram summarizing the overall experimental
Genome edi3ng with the CRISPR-Cas9 system
 CRISPR-Cas9 Genome Edi3ng Bootcamp AHA Council on Func3onal Genomics and Transla3onal Biology Narrated video link: hfps://youtu.be/h18hmftybnq Genome edi3ng with the CRISPR-Cas9 system Kiran Musunuru,
CRISPR-Cas9 Genome Edi3ng Bootcamp AHA Council on Func3onal Genomics and Transla3onal Biology Narrated video link: hfps://youtu.be/h18hmftybnq Genome edi3ng with the CRISPR-Cas9 system Kiran Musunuru,
Supporting Information
 Supporting Information Nowakowski et al. 10.1073/pnas.1219385110 SI Materials and Methods Primary Antibodies. Primary antibodies used in this study were raised against BrdU (rat, 1:50; Abcam), cleaved
Supporting Information Nowakowski et al. 10.1073/pnas.1219385110 SI Materials and Methods Primary Antibodies. Primary antibodies used in this study were raised against BrdU (rat, 1:50; Abcam), cleaved
Transcriptional Regulation in Eukaryotes
 Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Transcriptional Regulation in Eukaryotes Concepts, Strategies, and Techniques Michael Carey Stephen T. Smale COLD SPRING HARBOR LABORATORY PRESS NEW YORK 2000 Cold Spring Harbor Laboratory Press, 0-87969-537-4
Post-expansion antibody delivery, after epitope-preserving homogenization.
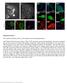 Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Supplementary Figure 1 Post-expansion antibody delivery, after epitope-preserving homogenization. (a, b) Wide-field fluorescence images of Thy1-YFP-expressing mouse brain hemisphere slice before expansion
Cell Stem Cell, volume 13 Supplemental Information
 Cell Stem Cell, volume 13 Supplemental Information Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells Ehsan Habibi, Arie B. Brinkman, Julia
Cell Stem Cell, volume 13 Supplemental Information Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells Ehsan Habibi, Arie B. Brinkman, Julia
Generation of ips-derived model cells for analyses of hair shaft differentiation
 Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
Supplementary Material Generation of ips-derived model cells for analyses of hair shaft differentiation Takumi Kido, Tomoatsu Horigome, Minori Uda, Naoki Adachi, Yohei Hirai Department of Biomedical Chemistry,
NAME TA SEC Problem Set 4 FRIDAY October 15, Answers to this problem set must be inserted into the box outside
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
Nature Methods: doi: /nmeth Supplementary Figure 1. Validation of RaPID with EDEN15
 Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
ChIP-seq/Functional Genomics/Epigenomics. CBSU/3CPG/CVG Next-Gen Sequencing Workshop. Josh Waterfall. March 31, 2010
 ChIP-seq/Functional Genomics/Epigenomics CBSU/3CPG/CVG Next-Gen Sequencing Workshop Josh Waterfall March 31, 2010 Outline Introduction to ChIP-seq Control data sets Peak/enriched region identification
ChIP-seq/Functional Genomics/Epigenomics CBSU/3CPG/CVG Next-Gen Sequencing Workshop Josh Waterfall March 31, 2010 Outline Introduction to ChIP-seq Control data sets Peak/enriched region identification
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Theoretical cloning project
 Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
Theoretical cloning project Needed to get credits Make it up yourself, don't copy Possible to do in groups of 2-4 students If you need help or an idea, ask! If you have no idea what to clone, I can give
SureSilencing sirna Array Technology Overview
 SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
SureSilencing sirna Array Technology Overview Pathway-Focused sirna-based RNA Interference Topics to be Covered Who is SuperArray? Brief Introduction to RNA Interference Challenges Facing RNA Interference
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
 A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
A idm-3 idm-3 B Physical distance (Mb) 4.6 4.86.6 8.4 C Chr.3 Recom. Rate (%) ATG 3.9.9.9 9.74 Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC
