Description of Supplementary Files. File name: Supplementary Information Description: Supplementary figures and supplementary tables.
|
|
|
- Sydney Smith
- 6 years ago
- Views:
Transcription
1 Description of Supplementary Files File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Supplementary Data 1 Description: Differential expression of annotated transcripts in list of transcripts that were differentially expressed in SKMM1 and H929 MM cell lines transduced with FAM46C WT -GFP compared to FAM46C mut -GFP transductions. Following columns with DESeq2 results are assigned to each comparison: 'basemean' average of the normalized count values, taken over mutant and wild type samples; 'log2foldchange' log2 fold change in the mutant comparing to wild type; 'lfcse' the standard error estimate for the log2 fold change estimate; 'stat' Wald significance test statistics; 'pvalue' - significance test p-value; 'padj' Benjamini-Hochberg adjusted p-value. File name: Supplementary Data 2 Description: List of transcripts polyadenylated in SKMM1 and H929 MM cell lines transduced with FAM46C WT -GFP compared to FAM46C mut -GFP transductions. Columns represent polyadenylation ratios calculated by dividing the levels of individual mrna in long poly(a) fractions (#5 and #6) relative to the short poly(a) fraction (#1) in wild-type FAM46C cells using relative levels observed in FAM46C mut cells. File name: Supplementary Data 3 Description: Mass spectrometry data from FAM46C immunoprecipitations. File name: Supplementary Data 4 Description: List of oligonucleotides used in this study. File name: Peer review file
2 Supplementary Figure 1. Multiple sequence alignments color-coded for amino acid conservation of FAM46 protein family representatives in humans performed by PRALINE. The scoring color scheme starts at 0 for the least conserved alignment position up to 10 for the most conserved alignment position. Invariant catalytic residues from highly conserved NTase domain ([DE]h[DE]h are shown in boxes (D90 and D92 in case of FAM46C).
3 Supplementary Figure 2. (a) Northern blot analysis of RL mrna from control HEK293 cells (lanes 4-9) after tethering of NHA-FAM46C WT (lanes 10-15) or NHA-FAM46C mut (lanes 16-21) which were fractionated based on the length of the poly(a) tails. Input RNAs are shown in lanes 1-3 (b) Northern blot analysis of RL mrna from control HEK293 cells (lanes 4-9) after tethering of NHA-FAM46D WT (lanes 10-15) or NHA-FAM46D mut (lanes 16-21) which were fractionated based on the length of the poly(a) tails. Input RNAs are shown in lanes 1-3. (c) Time course of actinomycin-d treatment. Northern blot detection of RL transcript from: control HEK293 cells (lanes 1-4), after tethering of NHA-FAM46C WT (lanes 5-8) and NHA- FAM46C mut (lanes 9-12). GAPDH mrna was used as loading controls for all northern blots. (d) RL mrna half-life quantifications shown as a plot with SD error bars (n=3).
4 Supplementary Figure 3. Expression of FAM46C WT induces multiple myeloma cell death. (a, b) Summary of flow cytometry analyses presented as bar graphs showing GFP expression levels and reduced viability of HEK293T (a) and RPMI8226 multiple myeloma cell line (b) depending on the amount of virus used for transduction and the time of expression analysis. Data are presented as percentage of cells ±SD (n=3). P values were calculated using 2-way ANOVA tests (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
5 Supplementary Figure 4. Different tags at the N and C termini of FAM46C protein do not affect the function of the protein. Summary of flow cytometry analyses presented as bar graphs showing reduced viability of multiple myeloma cell lines (H929, SKMM1, and RPMI8226) throughout the time course of FLAG-FAM46C, FAM46C-FLAG, GFP-FAM46C and NHA- FAM46C expression. Data are presented as percentage of cells ±SD (n=3). P values were calculated using 2-way ANOVA tests (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
6 Supplementary Figure 5. Expression of PAPOLA-GFP, POLS-GFP, and GLD2-GFP constructs had no major effect on multiple myeloma cell death. Summary of FACS analyses presented as bar graphs showing GFP expression level and reduced viability of SKMM1 multiple myeloma cell lines transduced with a lentivirus carrying PAPOLA, POLS, or GLD2. Data are represented as percentage of cells ± SD (n=3).
7 Supplementary Figure 6. FAM46C tethering enhanced expression of Renilla luciferase (RL) reporter in MM cells. SKMM1 (a) and H929 cells (b) expressing reporter genes of Renilla luciferase (RL) and Firefly luciferase (FL) or RL containing five boxb sites in its 3 -UTR (RL5Box) and FL were transduced with lentiviruses carrying FAM46C WT or FAM46C mut harboring the N-terminal λn boxb binding domain and HA-tag. Western blot detection of RL and FL proteins in non-transduced (a, b; lanes 1 and 4) and after NHA-FAM46C WT (a, b; lane 2 and 5) or NHA-FAM46C mut (a, b; lane 3 and 6) tethering. Expression of NHA-tagged FAM46C proteins were confirmed using an α-ha antibody. DBC1 and RPS5 served as a loading control. Asterisk indicates non-specific signals for HA and FLuc antibodies.
8 Supplementary Figure 7. Analysis of FAM46C substrates in MM cells. (a, b) Functional GO terms annotation clustering of significantly upregulated transcripts in FAM46C WT overexpressing SKMM1 (a) and H929 (b) cell lines. (c, d) Histogram representing the distribution of FAM46C-dependent polyadenylation rations monitored in fraction #5 (c) and #6 (d) in H929 cell lines. Modeled normal distribution is fitted as a red line to emphasize the outlying population of FAM46C polyadenylated transcripts. (e) The sequence composition of significantly overrepresented motifs (P value <10-12 ) in 3 UTRs of FAM46C substrates. The distance was calculated based on the Pearson Correlation Coefficient and an unweighted pair group method with arithmetic mean (UPGMA) tree-construction algorithms was used.
9 Supplementary Figure 8. SSR2 mrna, similarly to SSR4, is polyadenylated by FAM46C. (a) High resolution northern blot analysis of SSR2 transcript from MM cell lines H929 (lanes 1-3), SKMM1 (lanes 4-6) transduced with GFP (lanes 1, 4), FAM46C WT -GFP (lanes 2, 5), and FAM46C mut -GFP (lanes 3, 6). High-resolution northern blot analyses were performed using 4% denaturing PAGE gel. (b) High-resolution northern blot analysis of SSR2 transcripts from H929 (lanes 1-6) and SKMM1 cells (lanes 7-12) transduced with GFP (lanes 1, 2, 7, 8), FAM46C WT - GFP (lanes 3, 4, 9, 10), and FAM46C mut -GFP (lanes 5, 6, 11, 12) after RNase H treatment (lanes 2, 4, 6, 8, 10, 12) to remove the poly(a) tail in the presence of oligo(dt)25. Control reactions were carried out in the presence of oligo(dt)25 without RNase H (lanes 1, 3, 5, 7, 9, 11). (c) SSR4 is not the substrate for PAPOLA, POLS, and GLD2. High-resolution northern blot analysis of SSR4 transcripts from SKMM1 cells transduced with lentiviruses carrying PAPOLA (lane 2), POLS (lane 3), GLD2 (lane 4), and control cells (lane 1). RN7 RNA served as a loading control.
10 Supplementary Figure 9. (a-c) Tethering of PABPC1 to reporter RL mrna leads to increase of its steady state level. (a) Northern blot analysis of total RNA from HEK293 cells transfected with prl-5boxb and plasmids encoding NHA-BCCIPβ (lane 2) and NHA-PABPC1 (lane 3) using a probe against Renilla luciferase. Cells transfected with prl-5boxb only were used as controls (lane 1). Membrane methylene blue staining shown as a loading control. (b) Quantification of northern blots shown in panel a using Multigauge software. Bars represent mean values ± SD (n=3). (c) Western blot detection of NHA-BCCIPβ (lane 2) and NHA- PABPC1 (lane 3) with α-ha antibodies and reporter-encoded Renilla luciferase proteins using α-rl antibodies in control cells (lane 1). (d-f) Silencing of FAM46C interactors has no effect on its activity in tethering assays. (d) Western blot detection of BCCIP or PABPC1 proteins in control cells (lanes 1-3) and after sirna-mediated knockdowns (lanes 4-6). FAM46C tethering cells were transfected with prl-5boxb only (lanes 1, 4) or co-transfected additionally with plasmids encoding NHA-FAM46C WT (lanes 2, 5), and NHA-FAM46C mut (lanes 3, 6). (e) Western blot detection of NHA-FAM46C WT and NHA-FAM46C mut proteins using α-ha antibodies, Renilla luciferase proteins with an α-rl antibody in control HEK293 cells (lanes 1-3), or reduced levels of BCCIP (lanes 4-6) or PABPC1 (lanes 7-9). (f) Northern blot analysis of total RNA from control HEK293 cells (lanes 1-3) or with reduced levels of BCCIP (lanes 4-6) or PABPC1 (lanes 7-9) transfected with prl-5boxb only (lanes 1, 4, 7) or co-transfected additionally with plasmids encoding NHA-FAM46C WT (lanes 2, 5, 8) or NHA-FAM46C mut (lanes 3, 6, 9) using probes against Renilla luciferase. Membrane methylene blue staining is shown as a loading control.
11
12
13
14
15 Supplementary Figure 10. Unprocessed scans of selected blots and gels. (a) Scan of gel corresponding to Fig. 1b. (b) Blots corresponding to Fig. 1d. (c) Western blots corresponding to Fig. 3a, Fig. 3b, Fig. 3c. (d) Northern blots corresponding to Fig. 3e, 3h, 3i. (e) Western blots and membrane Ponceau S staining corresponding to Fig. 5a and Fig. 5b. (f) Northern blots corresponding to all panels of Fig. 8. (g) Western blots corresponding to Fig. 9b and Fig. 9c.
16 Supplementary Table 1. Cell lines used in this study. Multiple myeloma cell line Mutations in FAM46C gene SKMM1 H929 MM1.S RPMI8226 Homozygotic deletion c.519delt (p.i173fsx36) Hemizygotic insertion c insc (p.l93fsx15) Hemizygotic substitution c.808a>g (p.m270v) None Supplementary Table 2. Antibodies used in this study. Antibodies and Cell Isolation Kits Anti-Renilla Luciferase Anti-Firefly Luciferase Anti-BCCIP Anti-HA Anti-PABPC1 Anti-DBC1 Anti-FLAG Anti-GFP Anti-RPS5 Anti-GAPDH Anti-SSR4 Manufacturer and catalog number Millipore; Anti-Renilla Luciferase clone 5B11.2; MAB4400 Abcam; Anti-Firefly Luciferase antibody; ab21176 Abcam; Anti-BCCIP antibody; ab97577 Abcam; Anti-HA tag antibody [HA.C5]; ab18181 Abcam; Anti-PABP antibody; ab21060 Bethyl Lab; DBC1/p30 DBC Antibody; A A Invitrogen; DYKDDDDK Tag Polyclonal Antibody; PA1-984B Santa Cruz; GFP Antibody (B-2); sc-9996 Santa Cruz; Ribosomal Protein S5 (464-J); sc Novus Biologicals; anti-gapdh antibody; NB Abcam; Anti-Signal sequence receptor delta; ab58009 Anti-Histone H4 Anti-Histone H4, pan, clone ; Anti-FAM46C 1 Anti-FAM46C 2 Anti-FAM46C 3 Anti-FAM46C 4 Anti-GRP94 Abcam; Anti-FAM46C antibody; ab74754 Abcam; Anti-FAM46C antibody; ab Santa Cruz; FAM46C (D-14); sc Santa Cruz; FAM46C (P-12); sc Santa Cruz; GRP 94 (9G10); sc-32249
17 anti-cd20-alexa Fluor 700 Novus Biologicals; MS4A1/CD20 Antibody (AISB12); NBP AF700 Anti-CD138-PE DB; BD anti-cd45.2 Horizon V500 Anti-PSPC1 B Cell Isolation Kit, mouse CD138+ Plasma Cell Isolation Kit, mouse DB; anti-mouse CD45.2 (Clone 104); BD Abcam; Anti-PSPC1 antibody (ab104238) Miltenyi Biotec; Miltenyi Biotec; Supplementary Table 3. sirnas used in this study. Targeted gene Stealth Select RNAi sirna reference number Name used in this study BCCIP HSS sibccip_1 BCCIP HSS sibccip_2 BCCIP HSS sibccip_3 PABPC1 HSS sipabpc1_1 PABPC1 HSS sipabpc1_2 PABPC1 HSS sipabpc1_3 Supplementary Table 4. shrnas used in this study. construct Catalog or TRC Number plko.1-empty SHC001 (plko.1-puro Empty Vector) sh2 (hfam46c TRCN sh2) sh3 (hfam46c TRCN sh3) sh5 (hfam46c TRCN sh5) Sequence of insert No shrna Insert CCGGGCAGAATTTCAGCTGGTTAGACTCGAGTCTAACCA GCTGAAATTCTGCTTTTTTG CCGGGCCTAAATCTTGTTTACCTATCTCGAGATAGGTAAA CAAGATTTAGGCTTTTTTG CCGGGCTGAAGTTTGTCGACTCCATCTCGAGATGGAGTCG ACAAACTTCAGCTTTTTTG
18 Supplementary Table 5. Genotypes of mice used in this study. Mouse lines FAM46C-/- mouse line Mutations in FAM46C gene Homozygotic indel c.261_271delins301nt 1 (p.g85fsx26) FAM46C-FLAG mouse line Homozygotic insertion c.1176ins24bp 2 (p.n391insdykddddk) 1 apparently this indel consists of c.261_271delttgcaaagatctggatc (g.16015_16061del) and insertion of translocated intronic duplication g.15373_15670 fragment: c.261_271delttgcaaagatctggatcinsaaccgacagggtcgatttctctctgcggt TAGAGGTCCTTCATTGTTTAGATGGCATGGTTAGTTTGCTATTCTTCATCTAAAGC AGTCCCCCTTTCCTCTCCCCCACATAGGTCAAGCACACCAGCTAGAGCACCCACA CAAAGCAAAGTAGTGCATTACCACCCTGCTGTAGAACGGAGCTTGTGCCTGGCTG GGTATTACTCTTGGTCGCAGCCGTGTGGAGCAGCAGGCACTGCTGACTGCTGTTT TGCAGGAAATTGTTCAGAGATGGGTCAGTGCCTCTGAACTGACTCGTCCGT 2 full sequence of the c.1176ins24bp: insgactacaaagacgatgacgacaag
Translation of HTT mrna with expanded CAG repeats is regulated by
 Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
Supplementary Information Translation of HTT mrna with expanded CAG repeats is regulated by the MID1-PP2A protein complex Sybille Krauß 1,*, Nadine Griesche 1, Ewa Jastrzebska 2,3, Changwei Chen 4, Désiree
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated
 Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Supplementary Figure 1. TRIM9 does not affect AP-1, NF-AT or ISRE activity. (a,b) At 24h post-transfection with TRIM9 or vector and indicated reporter luciferase constructs, HEK293T cells were stimulated
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Online Supplementary Information
 Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Online Supplementary Information NLRP4 negatively regulates type I interferon signaling by targeting TBK1 for degradation via E3 ubiquitin ligase DTX4 Jun Cui 1,4,6,7, Yinyin Li 1,5,6,7, Liang Zhu 1, Dan
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles
 Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Thermo Scientific Dharmacon SMARTvector 2.0 Lentiviral shrna Particles Long-term gene silencing shrna-specific design algorithm High titer, purified particles Thermo Scientific Dharmacon SMARTvector shrna
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency
 Molecular Cell, Volume 52 Supplemental Information SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency Kengo Homma, Takao Fujisawa, Naomi Tsuburaya, Namiko
Molecular Cell, Volume 52 Supplemental Information SOD1 as a Molecular Switch for Initiating the Homeostatic ER Stress Response under Zinc Deficiency Kengo Homma, Takao Fujisawa, Naomi Tsuburaya, Namiko
TE5 KYSE510 TE7 KYSE70 KYSE140
 TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
TE5 KYSE5 TT KYSE7 KYSE4 Supplementary Figure. Hockey stick plots showing input normalized, rank ordered H3K7ac signals for the candidate SE-associated lncrnas in this study. Rpm Rpm Rpm Chip-seq H3K7ac
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Nature Methods: doi: /nmeth Supplementary Figure 1. Validation of RaPID with EDEN15
 Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
Supplementary Figure 1 Validation of RaPID with EDEN15 (a) Full Western Blot of conventional biotinylated RNA pulldown with EDEN15 and scrambled control (n=3 biologically independent experiments, representative
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
ENCODE DCC Antibody Validation Document Date of Submission 09/12/12 Name: Trupti Kawli Email: trupti@stanford.edu Lab Snyder Antibody Name: SREBP1 (sc-8984) Target: SREBP1 Company/ Source: Santa Cruz Biotechnology
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
doi:10.1038/nature09732 Supplementary Figure 1: Depletion of Fbw7 results in elevated Mcl-1 abundance. a, Total thymocytes from 8-wk-old Lck-Cre/Fbw7 +/fl (Control) or Lck-Cre/Fbw7 fl/fl (Fbw7 KO) mice
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
www.sciencemag.org/cgi/content/full/1137999/dc1 Supporting Online Material for Disrupting the Pairing Between let-7 and Enhances Oncogenic Transformation Christine Mayr, Michael T. Hemann, David P. Bartel*
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Khaled_Fig. S MITF TYROSINASE R²= MITF PDE4D R²= Variance from mean mrna expression
 Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Khaled_Fig. S Variance from mean mrna expression.8 2 MITF.6 TYROSINASE.4 R²=.73.2.8.6.4.2.8.6.4.2.8.6.4.2 MALME3M SKMEL28 UACC257 MITF PDE4D R²=.63 4.5 5 MITF 3.5 4 PDE4B 2.5 3 R²=.2.5 2.5.8.6.4.2.8.6.4.2
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing
 Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
Supplementary Information Design of small molecule-responsive micrornas based on structural requirements for Drosha processing Chase L. Beisel, Yvonne Y. Chen, Stephanie J. Culler, Kevin G. Hoff, & Christina
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Supplementary Figures Supplementary Figure 1 Experimental schema for the identification of circular RNAs in six normal tissues and seven cancerous tissues. Supplementary Fiure 2 Comparison of human circrnas
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues
 Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
Table I. Primers used in quantitative real-time PCR for detecting gene expressions in mouse liver tissues Gene name Forward primer 5' to 3' Gene name Reverse primer 5' to 3' CC_F TGCCCTGTTGGGGTTG CC_R
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
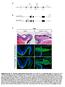 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Supplementary Information
 Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Journal : Nature Biotechnology Supplementary Information Targeted genome engineering in human cells with RNA-guided endonucleases Seung Woo Cho, Sojung Kim, Jong Min Kim, and Jin-Soo Kim* National Creative
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Galina Gabriely, Ph.D. BWH/HMS
 Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
Galina Gabriely, Ph.D. BWH/HMS Email: ggabriely@rics.bwh.harvard.edu Outline: microrna overview microrna expression analysis microrna functional analysis microrna (mirna) Characteristics mirnas discovered
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
affects the development of newborn neurons Brain Mind Institute and School of Life Sciences, Ecole Polytechnique Fédérale de
 Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Shedding of neurexin 3β ectodomain by ADAM10 releases a soluble fragment that affects the development of newborn neurons Erika Borcel* a, Magda Palczynska* a, Marine Krzisch b, Mitko Dimitrov a, Giorgio
Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive
 Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Supplemental Data Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by hemeresponsive gene-2 Caiyong Chen 1, Tamika K. Samuel 1, Michael Krause 2, Harry A. Dailey 3, and Iqbal
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Nature Genetics: doi: /ng Supplementary Figure 1. DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome.
 Supplementary Figure 1 DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome. (a,b) Quantitative RT-PCR analysis of CD4 + T cells isolated from the peripheral blood of individuals
Supplementary Figure 1 DNMT3A and TP53 expression analysis in samples from patients with Sézary syndrome. (a,b) Quantitative RT-PCR analysis of CD4 + T cells isolated from the peripheral blood of individuals
Supplemental Information. The TRAIL-Induced Cancer Secretome. Promotes a Tumor-Supportive Immune. Microenvironment via CCR2
 Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Molecular Cell, Volume 65 Supplemental Information The TRAIL-Induced Cancer Secretome Promotes a Tumor-Supportive Immune Microenvironment via CCR2 Torsten Hartwig, Antonella Montinaro, Silvia von Karstedt,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Isolation, culture, and transfection of primary mammary epithelial organoids
 Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
Supplementary Experimental Procedures Isolation, culture, and transfection of primary mammary epithelial organoids Primary mammary epithelial organoids were prepared from 8-week-old CD1 mice (Charles River)
CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611
 Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
Data Sheet CRE/CREB Reporter Assay Kit camp/pka Cell Signaling Pathway Catalog #: 60611 Background The main role of the camp response element, or CRE, is mediating the effects of Protein Kinase A (PKA)
MISSION shrna Library: Next Generation RNA Interference
 Page 1 of 6 Page 1 of 6 Return to Web Version MISSION shrna Library: Next Generation RNA Interference By: Stephanie Uder, Henry George, Betsy Boedeker, LSI Volume 6 Article 2 Introduction The technology
Page 1 of 6 Page 1 of 6 Return to Web Version MISSION shrna Library: Next Generation RNA Interference By: Stephanie Uder, Henry George, Betsy Boedeker, LSI Volume 6 Article 2 Introduction The technology
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Introduction to RNA-Seq. David Wood Winter School in Mathematics and Computational Biology July 1, 2013
 Introduction to RNA-Seq David Wood Winter School in Mathematics and Computational Biology July 1, 2013 Abundance RNA is... Diverse Dynamic Central DNA rrna Epigenetics trna RNA mrna Time Protein Abundance
Introduction to RNA-Seq David Wood Winter School in Mathematics and Computational Biology July 1, 2013 Abundance RNA is... Diverse Dynamic Central DNA rrna Epigenetics trna RNA mrna Time Protein Abundance
Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More. Ed Davis, Ph.D.
 TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss
 SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
SUPPLEMENTARY INFORMATION Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss Yunjong Lee, Senthilkumar S. Karuppagounder, Joo-Ho Shin, Yun-Il Lee, Han Seok Ko, Debbie Swing,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
B Vehicle 1V270 (35 μg) 1V270 (100 μg) Days post tumor implantation. Vehicle 100μg 1V270 biweekly 100μg 1V270 daily
 Supplemental Figure 1 A 1 1V7 (8 μg) 1 1V7 (16 μg) 8 6 4 B 1 1 8 6 4 1V7 (35 μg) 1V7 (1 μg) 5 1 15 5 1 15 C Biweekly 8 11 14 17 Days Implant SCC7 cells Daily 8 9 1 11 1 Implant SCC7 cells 1V7 i.t. treatment
Supplemental Figure 1 A 1 1V7 (8 μg) 1 1V7 (16 μg) 8 6 4 B 1 1 8 6 4 1V7 (35 μg) 1V7 (1 μg) 5 1 15 5 1 15 C Biweekly 8 11 14 17 Days Implant SCC7 cells Daily 8 9 1 11 1 Implant SCC7 cells 1V7 i.t. treatment
Supplemental Information. Mitophagy Controls the Activities. of Tumor Suppressor p53. to Regulate Hepatic Cancer Stem Cells
 Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
Molecular Cell, Volume 68 Supplemental Information Mitophagy Controls the Activities of Tumor Suppressor to Regulate Hepatic Cancer Stem Cells Kai Liu, Jiyoung Lee, Ja Yeon Kim, Linya Wang, Yongjun Tian,
X2-C/X1-Y X2-C/VCAM-Y. FRET efficiency. Ratio YFP/CFP
 FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
FRET efficiency.7.6..4.3.2 X2-C/X1-Y X2-C/VCAM-Y.1 1 2 3 Ratio YFP/CFP Supplemental Data 1. Analysis of / heterodimers in live cells using FRET. FRET saturation curves were obtained using cells transiently
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
microrna-122 stimulates translation of Hepatitis C Virus RNA
 Supplementary information for: microrna-122 stimulates translation of Hepatitis C Virus RNA Jura Inga Henke 1,5, Dagmar Goergen 1,5, Junfeng Zheng 3, Yutong Song 4, Christian G. Schüttler 2, Carmen Fehr
Supplementary information for: microrna-122 stimulates translation of Hepatitis C Virus RNA Jura Inga Henke 1,5, Dagmar Goergen 1,5, Junfeng Zheng 3, Yutong Song 4, Christian G. Schüttler 2, Carmen Fehr
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA
 sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
sirna Transfection Into Primary Neurons Using Fuse-It-siRNA This Application Note describes a protocol for sirna transfection into sensitive, primary cortical neurons using Fuse-It-siRNA. This innovative
Supplemental Information
 Supplemental Information Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene Chuan Shen, Rameen Beroukhim, Steven E. Schumacher, Jing Zhou, Michelle Chang, Sabina Signoretti,
Supplemental Information Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene Chuan Shen, Rameen Beroukhim, Steven E. Schumacher, Jing Zhou, Michelle Chang, Sabina Signoretti,
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and
 Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
Supplementary Figure 1: Derivation and characterization of RN ips cell lines. (a) RN ips cells maintain expression of pluripotency markers OCT4 and SSEA4 after 10 passages in mtesr 1 medium. (b) Schematic
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p53 activity
 IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
IKK is a therapeutic target in KRAS-induced lung cancer with disrupted p5 activity H6 5 5 H58 A59 H6 H58 A59 anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα anti-ikkβ anti-panras anti-gapdh anti-ikkα
M X 500 µl. M X 1000 µl
 GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
GeneGlide TM sirna Transfection Reagent (Catalog # M1081-300, -500, -1000; Store at 4 C) I. Introduction: BioVision s GeneGlide TM sirna Transfection reagent is a cationic proprietary polymer/lipid formulation,
SUPPLEMENTAL MATERIAL. Supplemental Methods. Cumate solution was from System Biosciences. Human complement C1q and complement
 SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
SUPPLEMENTAL MATERIAL Supplemental Methods Reagents Cumate solution was from System Biosciences. Human complement Cq and complement C-esterase inhibitor (C-INH) were from Calbiochem. C-INH (Berinert) for
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
GTTCGGGTTCC TTTTGAGCAG
 Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figures Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Wang C, Zhu H, Jin L, Chen T, Wang L, Kang H, Hong Z, Zhang Z. 5 UTR CDS 3 UTR TCTCAACCATCCTTTGTCTGCTTCCGCCGCATGGGTGAGGTCATTTTGTCTAGATGACGTGCAATTTACAATGA
Supplementary Figures and Legends
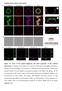 Supplementary Figures and Legends Figure S1. Tests of the optical alignment and focal properties of the confocal microscope. (A) Images of the optical cross-section of fluorescent microspheres differing
Supplementary Figures and Legends Figure S1. Tests of the optical alignment and focal properties of the confocal microscope. (A) Images of the optical cross-section of fluorescent microspheres differing
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc RGLG Hormonal treatment H2O B RGLG µm ABA µm ACC µm GA Time (hours) µm µm MJ µm IA
 Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Supplemental Data. Wu et al. (2). Plant Cell..5/tpc..4. A B Supplemental Figure. Immunoblot analysis verifies the expression of the AD-PP2C and BD-RGLG proteins in the Y2H assay. Total proteins were extracted
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Material
 Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
Fig S1. Interactions of OsDof24 with the OsC4PPDK promoter in rice protoplasts (a) Loss-of-function analysis of the OsC4PPDK promoter Effects of OsDof24 overexpression and the mapping of OsDof24-binding
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
CH 17 :From Gene to Protein
 CH 17 :From Gene to Protein Defining a gene gene gene Defining a gene is problematic because one gene can code for several protein products, some genes code only for RNA, two genes can overlap, and there
CH 17 :From Gene to Protein Defining a gene gene gene Defining a gene is problematic because one gene can code for several protein products, some genes code only for RNA, two genes can overlap, and there
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression
 Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Yue Wang, Zhenyu Xu, Junfeng Jiang, Chen Xu, Jiuhong Kang, Lei Xiao, Minjuan Wu, Jun Xiong, Xiaocan Guo, and Houqi Liu
 Developmental Cell, Volume 25 Supplemental Information Endogenous mirna Sponge lincrna-ror Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal Yue Wang, Zhenyu Xu, Junfeng Jiang,
Developmental Cell, Volume 25 Supplemental Information Endogenous mirna Sponge lincrna-ror Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal Yue Wang, Zhenyu Xu, Junfeng Jiang,
Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and
 Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supporting Information
 Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
Supporting Information Horie et al. 10.1073/pnas.1008499107 SI Materials and Methods ell ulture and Reagents. THP-1 cells were obtained from the American Type ell ollection. THP-1 cells were transformed
measuring gene expression December 5, 2017
 measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
measuring gene expression December 5, 2017 transcription a usually short-lived RNA copy of the DNA is created through transcription RNA is exported to the cytoplasm to encode proteins some types of RNA
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Supplemental Data Supplementary Figure Legends and Scheme Figure S1.
 Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Supplemental Data Supplementary Figure Legends and Scheme Figure S1. UTK1 inhibits the second EGF-induced wave of lamellipodia formation in TT cells. A and B, EGF-induced lamellipodia formation in TT cells,
Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna
 Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
Supplemental Materials and Methods Short hairpin RNA (shrna) against MMP14. Lentiviral plasmids containing shrna (Mission shrna, Sigma) against mouse MMP14 were transfected into HEK293 cells using FuGene6
SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and
 SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and first strand cdna was synthesized using 1 μg of RNA and
SUPPLEMENTAL METHODS Reverse transcriptase PCR (RT-PCR) and quantitative real-time PCR (Q-RT-PCR) RNA was isolated with TRIZOL (Invitrogen), and first strand cdna was synthesized using 1 μg of RNA and
Supplemental Data. Sun et al. Plant Cell. (2012) /tpc FLS2 -cmyc -GFP FLS2 -HA. FLS2-FLAG FLS2 -HA BAK1-HA flg22 (10mM)
 Supplemental Data. Sun et al. Plant ell. ()..5/tpc..9599 A FLS-FLA FLS -HA BAK-HA flg (mm) - B FLS -cmyc -FP FLS -HA IP antibody cmyc FLA FP cmyc IP ; WB: α-ha IP; WB: α-ha IP: α-fla; WB: α-ha IP ; WB:
Supplemental Data. Sun et al. Plant ell. ()..5/tpc..9599 A FLS-FLA FLS -HA BAK-HA flg (mm) - B FLS -cmyc -FP FLS -HA IP antibody cmyc FLA FP cmyc IP ; WB: α-ha IP; WB: α-ha IP: α-fla; WB: α-ha IP ; WB:
Color-Switch CRE Reporter Stable Cell Line
 Color-Switch CRE Reporter Stable Cell Line Catalog Number SC018-Bsd SC018-Neo SC018-Puro Product Name / Description CRE reporter cell line (Bsd): HEK293-loxP-GFP- RFP (Bsd). Cell line expresses "LoxP-GFP-stop-
Color-Switch CRE Reporter Stable Cell Line Catalog Number SC018-Bsd SC018-Neo SC018-Puro Product Name / Description CRE reporter cell line (Bsd): HEK293-loxP-GFP- RFP (Bsd). Cell line expresses "LoxP-GFP-stop-
1 Name. 1. (3 pts) What is apoptosis and how does it differ from necrosis? Which is more likely to trigger inflammation?
 1 Name MCB 150 Midterm Eam #1 (100 points total) Please write your full name on each page of the eam!! The eam consists of 17 questions (6 pages). Each has a different point count as indicated. Please
1 Name MCB 150 Midterm Eam #1 (100 points total) Please write your full name on each page of the eam!! The eam consists of 17 questions (6 pages). Each has a different point count as indicated. Please
